Numerous fundamental functions of ribonucleic acid (RNA) are involved in basic biological processes. It is imperative that we deepen our comprehension of the structures and activities of RNA, as it has emerged as an intriguing target for pharmaceuticals in recent times. Sequencing technologies have yielded vast amounts of unlabeled RNA data throughout the years, concealing valuable information and untapped potential. Inspired by the achievements of protein language models, scientists present the RiboNucleic Acid Language Model (RiNALMo) in an effort to decipher the RNA’s secret code. With 650 million parameters pre-trained on 36 million non-coding RNA sequences from many publicly available databases, RiNALMo is the largest RNA language model to date. It is possible for RiNALMo to uncover buried information and retrieve the underlying structural information that is subtly present in RNA sequences. RiNALMo achieves state-of-the-art performance in a number of downstream activities. Notably, the researchers demonstrate that its capacity for generalization can outperform previous deep learning techniques for secondary structure prediction in terms of its ability to generalize to unidentified RNA families.
Introduction
Massive text corpora have been used to train large language models (LLMs), which have shown impressive performance on a variety of natural language generation and processing tasks. The study of language models has expanded in recent years to include data from biology and fields other than natural language processing. Large-scale sequenced protein data served as a training platform for protein language models, which have since proven to be invaluable tools for tasks involving the generative and structural prediction of proteins.
There are currently just two single-input-sequence RNA foundation models that have been evaluated on various structural and function prediction tasks: RNA-FM and UniRNA.
Based on the initial implementation proposed by Vaswani et al. (2017), RNA-FM is a transformer encoder with 100 million parameters that were trained solely on 23.7 million non-coding RNAs (ncRNAs) from the RNAcentral database (RNAcentral Consortium, 2020). With an architecture similar to the ESM protein language model, Wang et al. (2023) pre-trained an ensemble of language models with 25–400 million parameters using a significantly bigger dataset of 1 billion sequences. In contrast to Uni-RNA, RNA-FM is an open-source RNA foundation model.
Role of RNA in Biological Functions
Important biological activities such as transcription, cell signaling, chromatin remodeling, and genome imprinting depend on ribonucleic acids. Similar to proteins, RNAs have lately gained attention as promising targets for drugs because of their structural similarities and the ways in which they interact with other biological components. However, RNA-related problems have received far less attention when applying language models. This is partially due to the lack of relevant data and structures, as well as the fact that RNA-related problems are typically more challenging than protein-related problems.
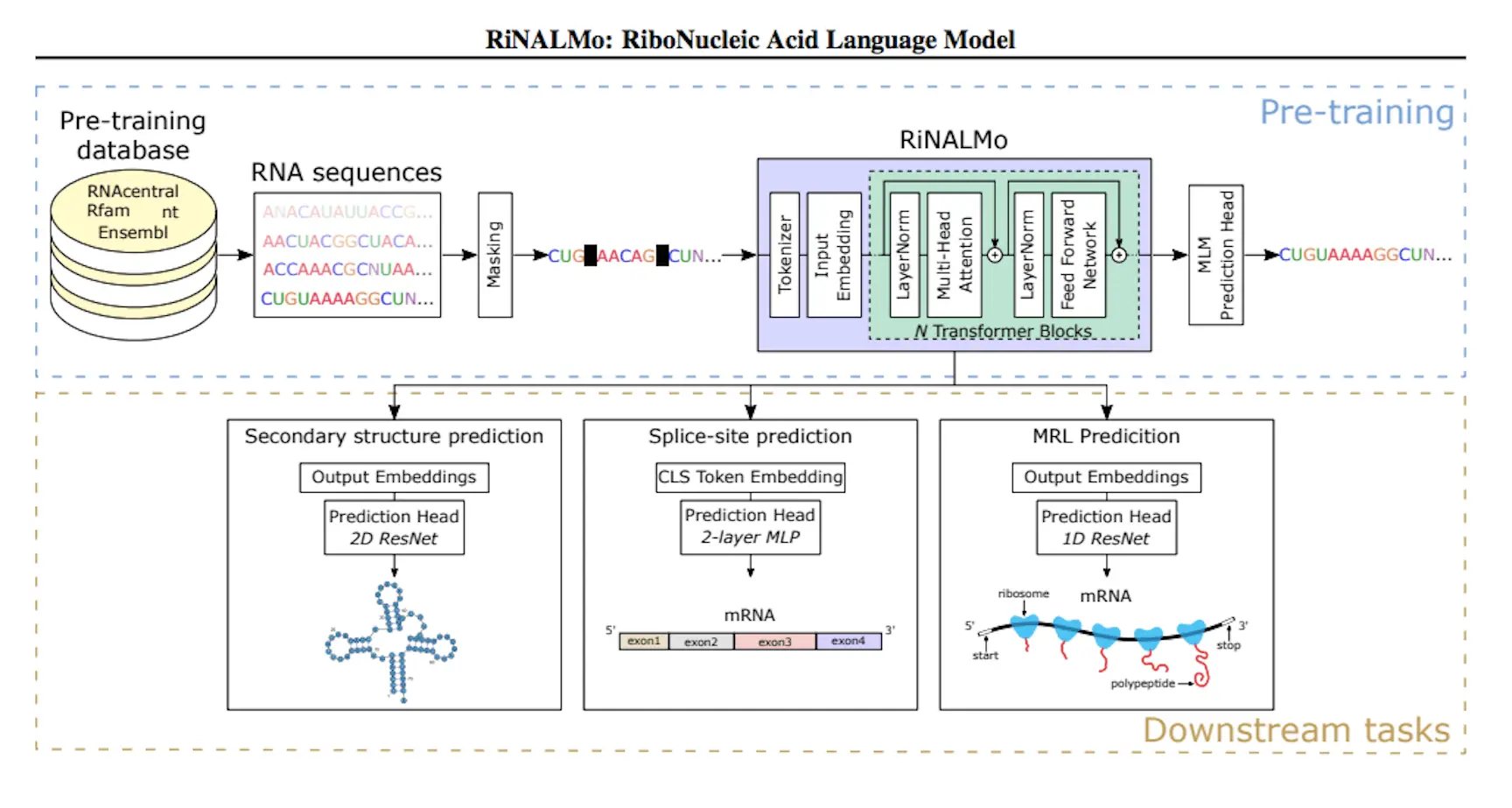
Understanding RiNALMo
Using 36 million ncRNA sequences and RNA databases, RiNALMo is a revolutionary RNA language model. It is a 650 million-parameter BERT-style transformer encoder that has been enhanced by contemporary methods such as FlashAttention-2, SwiGLU activation function, and rotary positional embedding (RoPE). During pre-training, RiNALMo uncovers hidden information and records underlying structural details at the single-nucleotide level in sequences. Performance on a variety of downstream structural and functional RNA tasks is enhanced by its output embeddings, especially when it comes to secondary structure prediction of RNA families that are not present in the training dataset.
3 Key Features of this Study
- The paper presents RiNALMo, the largest RNA language model to date with 650 million parameters that can fully utilize the potential of a massive number of publicly available unannotated RNA sequences.
- According to the study, existing deep learning techniques for secondary structure prediction have difficulty performing well on RNA families that are not present in the training dataset. However, RiNALMo’s generalization capability can solve this issue.
- Extensive studies on a variety of RNA structural and functional downstream tasks were carried out by the researchers, and the majority of datasets demonstrate that RiNALMo works better than existing RNA language models and deep learning techniques.
Methodology
Researchers examined and refined RiNALMo using two secondary structure prediction tasks. RNAs belonging to the same family were present in both the training and test datasets for the first task, which involved predicting the intra-family secondary structure. The findings demonstrated that when used in conjunction with an appropriate prediction head, fine-tuned RiNALMo’s output representation can achieve state-of-the-art performance by embedding crucial structure information about the input sequence. In the second task, which involved predicting an inter-family secondary structure, an RNA family from the test dataset had to be excluded from the training dataset. Previous research has demonstrated that deep learning techniques have poor cross-RNA family generalization. However, RiNALMo has shown that RNA language models may generalize well to unknown RNA families, outperforming both thermodynamics-based and deep-learning techniques. This illustrates RiNALMo’s exceptional capacity for generalization in secondary structure prediction challenges.
RiNALMo in Secondary Structure Prediction
RNAs generate complex structures that are essential to their stability and functionality. Tools for predicting tertiary structures require secondary structure information. Thermodynamic models are used by popular secondary structure prediction algorithms to determine which structures have the lowest free energy. Deep learning techniques and probabilistic approaches such as CONTRAfold have been developed as substitutes. However, these techniques frequently have difficulty generalizing well to novel RNA families, which limits their applicability. Notwithstanding these drawbacks, it is essential to apply these approaches to RNA structures in order to comprehend and forecast their characteristics.
Using binary cross-entropy loss, the RiNALMo model was trained on a binary classification challenge. Every nucleotide pair is categorized under the model as either paired or unpaired. The language model is coupled to a bottleneck residual neural network (ResNet), which is fed nucleotide pair representations produced by outer concatenating RiNALMo’s output with itself. The model was contrasted with well-known deep learning techniques for secondary structure prediction, including UFold, MXfold2, and RNA-FM. All models, with the exception of SPOT-RNA, which was refined on a smaller dataset taken from the PDB, were trained using the identical training dataset (TR0). RiNALMo performed noticeably better than the other methods.
The purpose of the study was to assess how well the RNA-FM model RiNALMo generalizes across various RNA families. 3865 RNAs from nine families made up the dataset, which was split nine times, with a different family being assessed for each split. The generalization capacity of the model was evaluated against several RNA-FM tools, including UFold, MXFold2, CONTRAfold, and RNAstructure. On each split, additional deep-learning models were trained independently while the language models were adjusted. The EternaBench dataset was used to train the EternaFold parameters for CONTRAfold secondary structure prediction.
In eight of the nine families, the model RiNALMo performs better than RNA structure, outperforming other models. It obtains the highest F1 score on all other families but fails to generalize on telomerase RNAs. The work displays the sequence embeddings for all RNAs in the dataset using RiNALMo, and it shows that telomerase RNAs are grouped together, while there isn’t a distinct separation from SRP RNAs. The dataset’s longest RNAs are telomerase RNAs, and UFold excels at processing them. Although the cause of RiNALMo’s inability to work with telomerase RNAs is still unknown, this issue will be addressed in the future.
Conclusion
An enormous number of ncRNA sequences from different databases, including RNAcentral and Rfam, were used to pre-train the RNA language model RiNALMo. Better generalization capabilities are the consequence of the data’s meticulous curation to guarantee sequence variety. The output representations of RiNALMo include details regarding RNA families and RNAs with comparable structures. On downstream tasks, the expressiveness and capacity of the model to extract information about hidden structures can be evaluated. In order to determine whether RiNALMo’s expressive output embeddings and generalization capabilities enhance current or novel prediction tools, future work will concentrate on applying RiNALMo to additional structure- and function-related tasks, notably tertiary structure prediction tasks.
Moreover, on two downstream RNA jobs relevant to function, researchers improved RiNALMo. These two tasks have to do with the mRNA family, for which there were no examples in the pre-training dataset. The findings demonstrated that RiNALMo could once again effectively generalize and extract significant functional information from RNA sequences belonging to hitherto undiscovered families.
Article source: Reference Paper | RiNALMo code has been made publicly available on GitHub
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.