Researchers at EvolutionaryScale introduced ESM3, an AI-driven language model for the life sciences that enables us to create and program using the universal language of life. In the same way researchers create machinery, structures, and microchips, this breakthrough advances the field of biological engineering from the ground up. ESM3 is very receptive to biological alignment and can respond to intricate cues that combine its modalities. Researchers have used a chain of thought to induce ESM3 to produce fluorescent proteins. Researchers discovered a brilliant fluorescent protein among the generations they synthesized that was far (58% identity) from previously identified fluorescent proteins. Natural fluorescent proteins that are similarly distant from one another have evolved over 500 million years apart.
Introduction
Around 3.5 billion years ago, chemical events gave rise to life on Earth and the development of RNA, proteins, and DNA. Building blocks for proteins are derived from DNA by the ribosome, a molecular factory. Proteins are dynamic molecules that perform amazing tasks such as scaffolding, information processing systems, molecular engines, and photosynthetic apparatuses. Proteins are the basis for both health and sickness and are the building blocks of many life-saving drugs. Because the ribosome can be programmed to make proteins from RNA codes, biology is the most advanced technology ever invented. There are dozens to millions of these molecular factories in every cell on Earth. However, biology is written in a language researchers do not fully comprehend, so even highly developed computational tools only scratch the surface. Biology might be programmed by learning to read and write the code of life, which would replace trial and error with simulation and logic.
What is ESM3?
A groundbreaking generative model for biology, EvolutionaryScale integrates the structure, function, and sequencing of proteins throughout Earth’s natural diversity. ESM3 is trained on one of the most productive GPU clusters in the world, showcasing the state-of-the-art in terms of parameters, computing capacity, and data. ESM3, with more than 1×1024 FLOPS and 98B parameters, is the biological model with the highest compute intensity ever used. Larger models acquire additional emergent capabilities that smaller models do not have when their size in parameters, data, and computation rises.
In biology, generalist models trained on various data sets outperform specialized models, demonstrating this trend. Over the previous five years, the ESM team has investigated scaling in biology and discovered that as language models scale, they gain a knowledge of the fundamental concepts of biology. Over the previous five years, the ESM team has investigated scaling in biology and discovered that as language models scale, they uncover biological structure and function and grasp the fundamental concepts of biology. An order of magnitude larger than earlier models and naturally multimodal and generative, ESM3 is a milestone model in the ESM family.
Unlocking Secrets of Life with AI
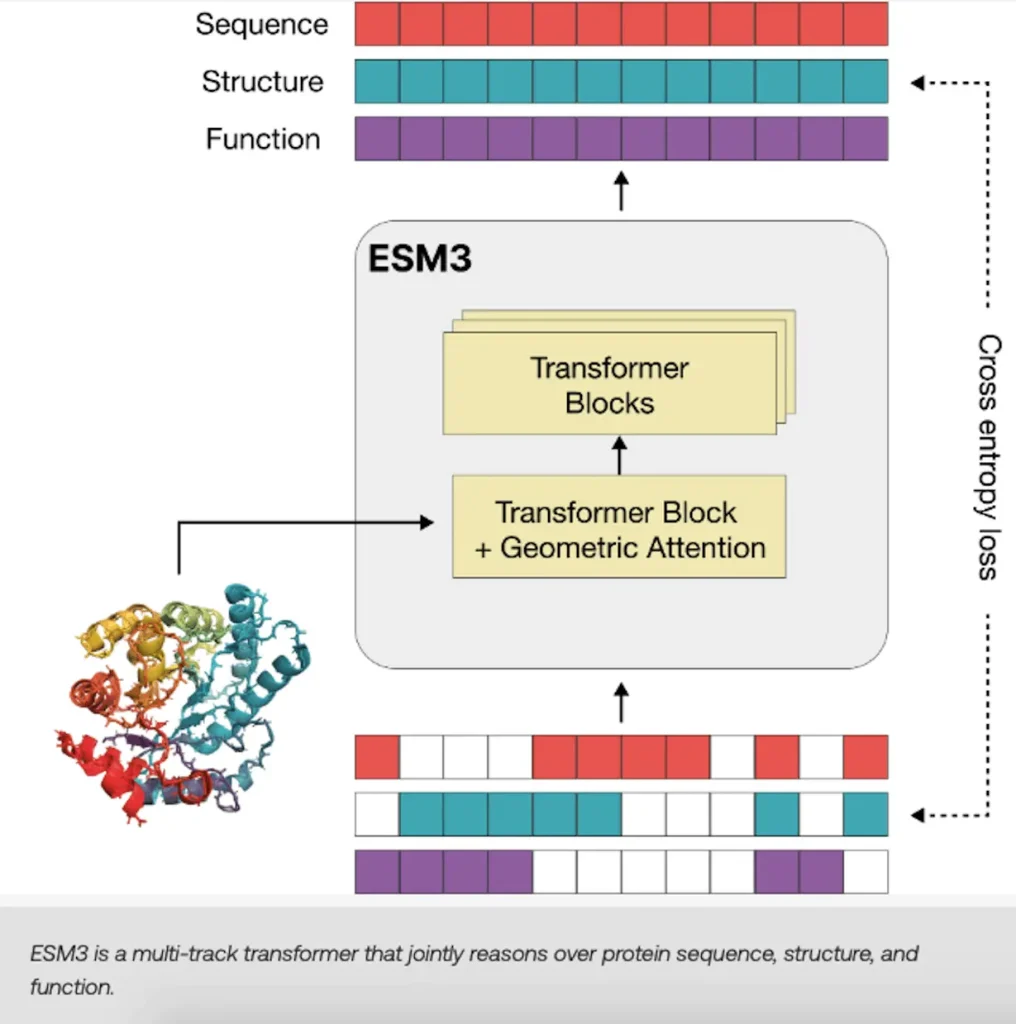
The language model ESM3 is intended to comprehend the basic biological characteristics of proteins, such as their sequencing, structure, and function. This is accomplished by writing three-dimensional structures that function as letter sequences after converting them into distinct alphabets. This opens the door to emergent generative capabilities in ESM3 by enabling training at scale. Within the same language model, the vocabulary of the model unites sequence, structure, and function. Using the masked language modeling aim, which natural language processing models influenced, the goal is to anticipate masked positions. When ESM3 is grown across billions of proteins and parameters, it can simulate evolution because it gains a profound understanding of the relationship between sequence, structure, and function across evolutionary-scale data.
With the generative model ESM3, researchers studying biology may now create new proteins with previously unheard-of control. During training, this model—which can produce proteins in all three modalities—is masked and makes predictions. By interacting with ESM3, scientists can produce proteins for use in a variety of fields, such as biology, medicine, and clean energy. Sequence, structure, and function can all be combined in the model to suggest a possible scaffold for PETase, an enzyme that breaks down PET and is a target for protein engineers looking to break down plastic waste. The multimodal reasoning capability of ESM3 enables scientists to design proteins with previously unheard-of control.
Scaling Up to Unlock Emergent Capabilities
When scaled up, ESM3 can perform better at solving difficult protein design problems like atomic coordination. Its achievement of atomic-level accuracy in structure formation makes it essential for the design of functional proteins. The scalability of ESM3’s solution to these jobs enables it to tackle more complex generating challenges. Like Reinforcement Learning from Human input (RLHF), used in LLMs, ESM3 also gets better with input. Giving itself feedback on the caliber of its generations and coordinating them with biological success enables ESM3 to develop itself.
500 Million Years of Evolution Simulated by AI
Beautiful proteins that can be found in nature in a variety of forms are fluorescent proteins, including green fluorescent protein (GFP). The protein’s ability to absorb and release light in many colors is made possible by a special mechanism called GFP folds. Many GFP variations have been found in nature and produced in labs by scientists. These proteins have evolved over eon of time; nature created the first fluorescent protein around 100 million years ago. The Nobel Prize was awarded for the discovery of GFP, which is now a commonly used biological tool.
The goal of ESM3 was to produce novel GFPs by chance by selecting from an enormous number of sequences and structures—many proteins fluorescence in an initial experiment that examined 96 generations. One protein, which matured over a week, was 50 times less brilliant than natural GFPs and was unlike any protein found in nature. Following the same reasoning, 96 proteins were produced, some of which were as brilliant as native GFPs. The brightest protein, esmGFP’s sequence, shared 58% similarities with the closest fluorescent protein in nature. Almost no sequence or structure can be created randomly due to their enormous quantity.
ESM3, like other protein language models, is not specifically made to function within evolutionary bounds. However, as an evolutionary simulator, they can discover how evolution proceeds through hypothetical proteins. The conventional analysis of evolution concerning esmGFP, a protein produced extraterrestrially, is contradictory. Nonetheless, the amount of time it would take for a protein to diverge from its closest sequence neighbor naturally can be estimated using tools from evolutionary biology. Hundreds of millions of years have separated naturally occurring GFPs with comparable sequence identities. esmGFP represents nearly 500 million years of natural evolution carried out using an evolutionary simulator, using an analysis akin to that carried out on a novel protein discovered in the natural world.
Conclusion
ESM3 is a tool that scientists can use to gain a fundamental knowledge of complicated biological systems and make novel scientific discoveries that alter the understanding of biology. Exploring protein design and synthetic biology boundaries enables scientists to create novel solutions for significant global issues. ESM3 is being developed for drug design applications, prioritizing beta access to the API based on the potential to further scientific understanding. The same tools scientists used to construct one of the most complicated proteins in nature will be utilized to help them design new medications. ESM3 is merely the beginning of the roadmap for programming biology; multimodal models that integrate across life scales and learn from biological data will be the main focus in the future. This will help humankind understand biology and program it to create better environments.
Article Source: Reference Article
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.
















[…] Simulating Evolution with EvolutionaryScale’s ESM3: A Leap Forward in Artificial Intelligence … […]