The development of single-cell profiling methods has transformed our understanding of the cellular and molecular processes underlying disease states and how they respond to therapy. However, existing computer methods cannot objectively recover known and new mechanistic insights at different levels of biological regulation. Here, researchers from the University of Western Australia and The University of Arizona introduce Decipher, a unique computational pipeline that determines the critical cellular and molecular events that drive disease and constructs integrated cell signaling networks from single-cell profiles in a context-specific, data-driven manner. The high-accuracy tool Decipher has been used to identify novel routes and context-specific effects in addition to recovering existing cytokine signaling pathways. It provides interpretable global cell-to-cell signaling maps with controllable interactions. Decipher can help identify new therapeutic targets and aid in developing new medications by deciphering signaling pathways and revealing cellular and molecular mechanisms.
Introduction
Systems Biology is a field that focuses on understanding the interactions between biological systems, particularly in the context of the development of complex multicellular life forms like humans. These systems carry out a number of tasks that are made possible by interactions between cells, including growth, differentiation, and development. Single-cell resolution profiling of biological systems is now possible thanks to recent developments in genomic profiling technology and computational techniques that have revolutionized our knowledge of these interactions. Unfortunately, because of the shortcomings of the tools at our disposal, our knowledge of the fundamental mechanisms by which these activities function in both health and sickness is still lacking. Although about 40 techniques have been developed to analyze cell-to-cell communication, the majority of these methods do not take the interactions’ long-term effects into account. Certain sophisticated techniques, such as gene regulatory networks (GRNs), simulate how ligand-receptor signaling affects the activation of downstream biological reactions, but their ability to discover new pathways is frequently limited by preexisting information.
Understanding Decipher
A revolutionary computational platform called Decipher is introduced in this work by researchers, which uses single-cell profiles to construct integrated cell signaling networks and reveals the essential cellular and molecular pathways that underlie disease processes and therapeutic responses. Researchers compare Decipher against other cutting-edge techniques and show how to apply it to single-cell profiling data from several experimental contexts. Researchers also show how Decipher can be used to derive mechanistic ideas from experimental data, emphasizing how this tool can be used to find new therapeutic targets and speed up the creation of new medications.
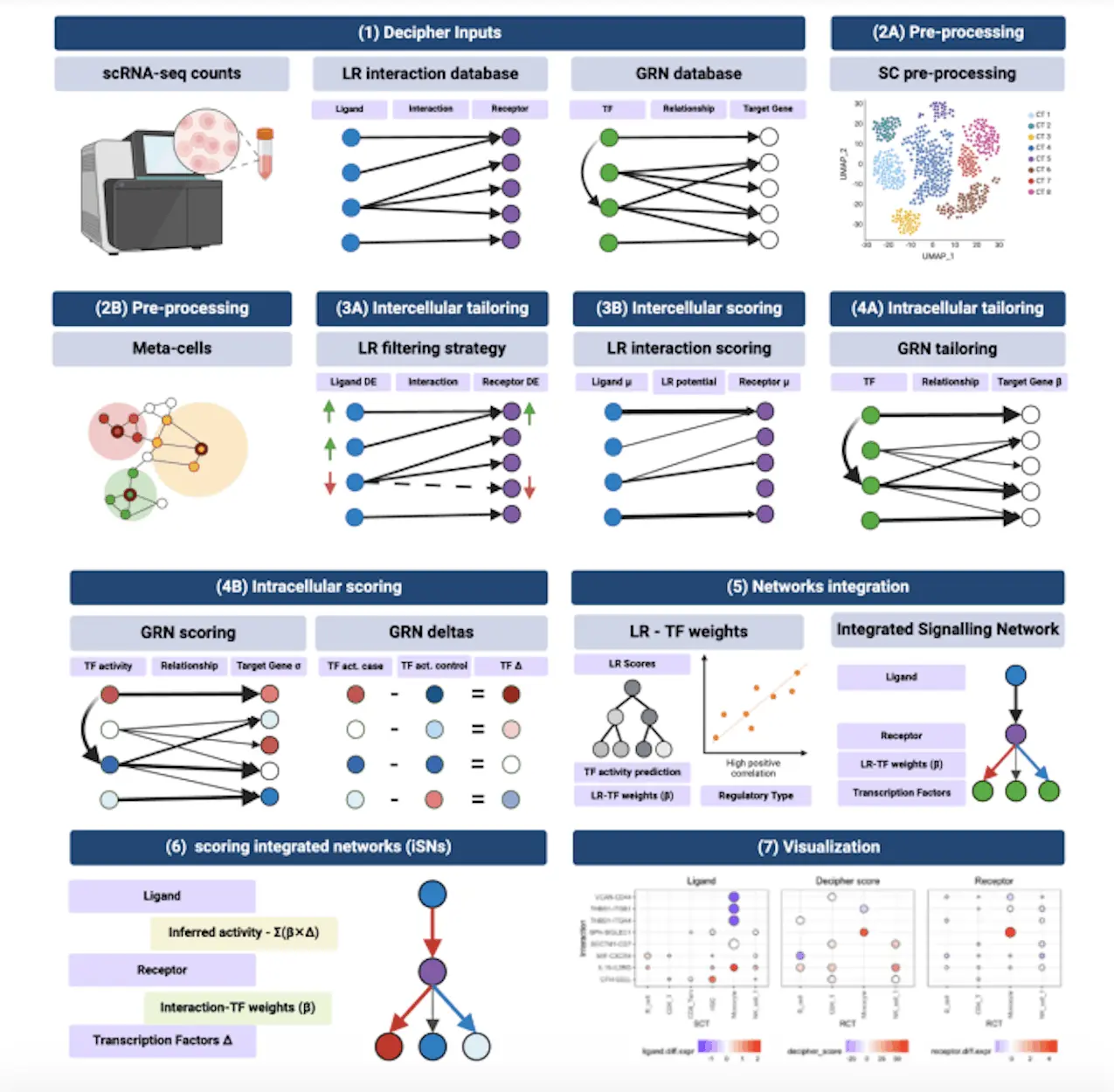
Decipher is a modular pipeline that links downstream intracellular responses mediated by transcription factors and target genes to intercellular signaling between ligand/receptor pairs. In order to customize, enhance, and extract mechanistic insights depending on the environment of interest, it incorporates biological networks. Decipher receives input single-cell RNA-seq profiles from two different experimental setups together with two previous knowledge networks: a Gene Regulatory Network (GRN) model and a database of reference Ligand-Receptor (LR) pairings. Depending on their experimental setup and environment, users employ the GRN from CellOracle and the LR pairings from connectomeDB2020, utilizing their selected prior knowledge resources.
Applications of Decipher
In addition to being able to recover existing signaling pathways accurately, Decipher generates a manageable collection of prioritized molecular pathways while retaining the ability to identify new routes. Furthermore, Decipher produces context- and cell-specific results according to its data-driven methodology. Together, these three elements are essential for bridging the gap that currently exists with current techniques and unlocking the critical cellular and molecular pathways that govern a biological system’s response to perturbations and/or disease processes. In addition, compared to other techniques like NicheNet and LIANA+, Decipher generates global cell-to-cell signaling maps that are simple to understand.
Decipher ultimately succeeded in regaining the mechanism responsible for a unique monocyte population enriched in genes triggered by interferons, which is notably more frequent after receiving the Pfizer-BioNTech COVID-19 mRNA vaccine as a secondary immunization.
Limitations
The scoring tool Decipher is limited by unbalanced intracellular activity. The Decipher scores, which are correlated linearly with intracellular activity, may be affected by the higher number of differentially active transcription factors (TFs) in some CTs. One possible workaround for this is to apply absolute scaling to TF activity and add a normalization factor, like in scSeqComm, where TF activity ∈ [0,1] and the normalization factor is the sum of the prior association scores for the TFs in a given pathway. A significant correlation between TF activity within each CT is expected, given the multi-collinearity and redundancy found in biological networks. It is recommended to use the new scoring schemes for TF activity that the bioinformatics community is actively developing. Decipher also does not consider feedback control or response dynamics, which can be addressed by extending Decipher to spatial transcriptomics data.
Conclusion
Mechanistic insights can be extracted through the comprehensive analysis of single-cell profiles provided by the modular pipeline Decipher. Decoding biological responses through ligand-receptor mediated TF-target gene networks, mapping cell-cell communication to a systems level, quantifying active LR pairs, and accounts for downstream intracellular responses. This tool is anticipated to speed up the process of finding new treatment targets for human illnesses.
Article Source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.