Although the field of protein engineering offers countless applications in chemistry, energy, and medicine, the process of designing new proteins with enhanced or unique functionalities is still labor-intensive, slow, and ineffective. The Self-driving Autonomous Machines for Protein Landscape Exploration (SAMPLE) platform for completely autonomous protein engineering is presented here by researchers from the University of Wisconsin–Madison. Automating and speeding up scientific discovery, self-driving labs have enormous potential in synthetic biology and protein engineering.
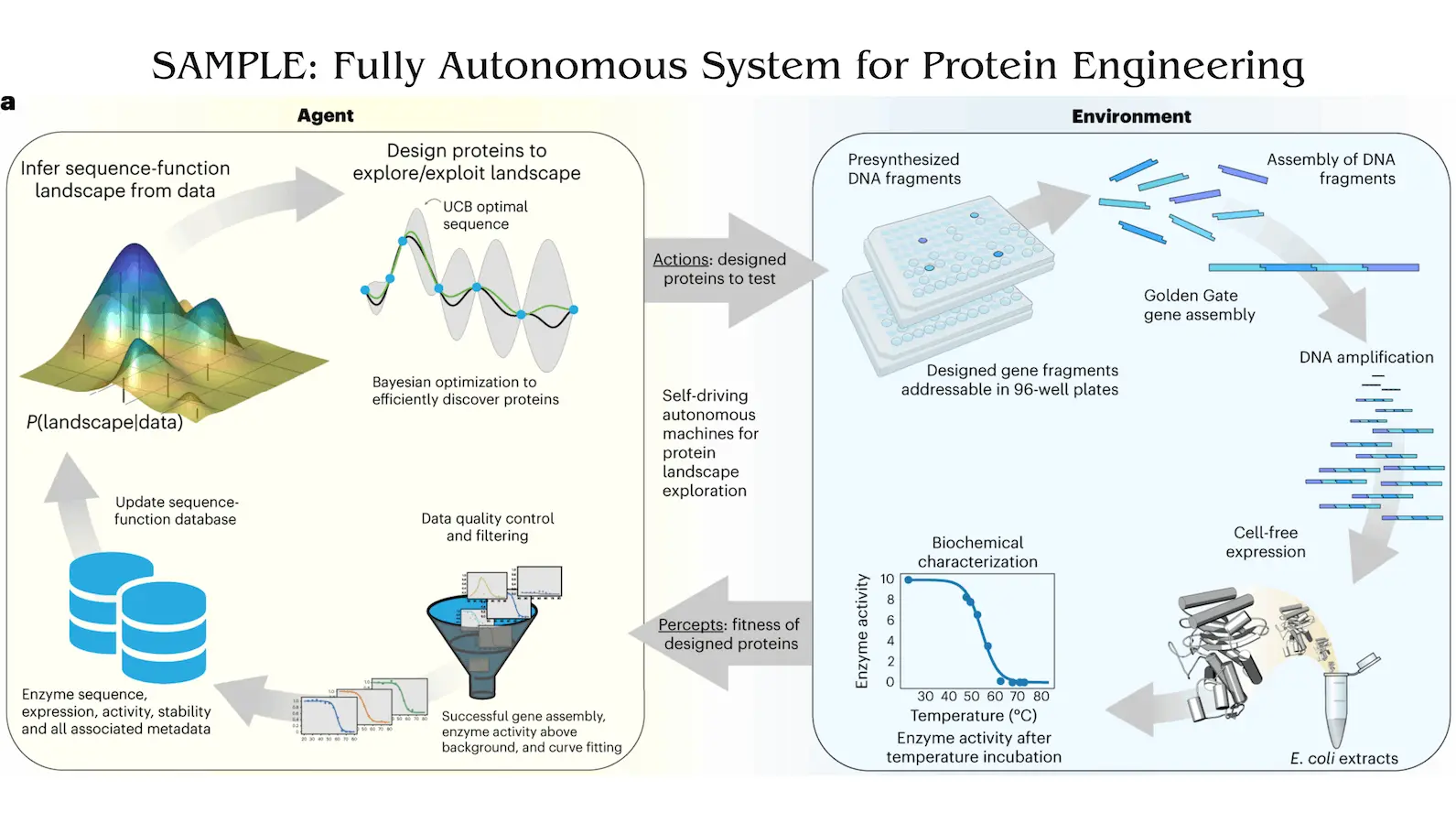
Driven by an intelligent agent, SAMPLE discovers correlations between the sequences of proteins and their functions. It then creates new proteins and delivers them to a completely automated robotic system, which tests them in an experiment and gives the agent feedback to help it better understand the system. In order to develop glycoside hydrolase enzymes with improved temperature endurance, the researchers utilize four SAMPLE agents. All four agents exhibit distinct variations in their search patterns, yet they rapidly come together around thermostable enzymes. Automating and speeding up scientific discovery, self-driving labs have enormous potential in synthetic biology and protein engineering.
Robotics and Automation
Synthetic biology is being revolutionized by robot scientists who combine automated reasoning, learning, and experimentation in self-driving labs. These intelligent systems can produce highly reproducible data, learn from a variety of data sources, make judgments in the face of uncertainty, and run nonstop. Gene identification, chemical synthesis techniques, and the development of novel materials such as adhesives, thin-film materials, photovoltaics, and photocatalysts have all benefited from their application. Complex biological phenotypes, high-dimensional genomic search fields, and labor-intensive manual processing processes present difficulties for these applications, too. Synthetic biology automated workflows can function without human input in certain cases, but they are not completely autonomous.
Understanding the Protein Fitness Landscape
The mapping from sequence to function is described by the protein fitness landscape, which is a topographical feature consisting of peaks, valleys, and ridges. Finding high-activity fitness peaks, or top-performing sequences, from an originally unknown sequence–function landscape is the goal of the SAMPLE agent. The agent actively probes its surroundings in order to obtain data and create an internal representation of the terrain. The agent has to divide its resources between exploitation—using existing knowledge of the landscape to find the best sequence configurations—and exploration—understanding the structure of the landscape. The protein engineering assignment for the agent is formulated as a Bayesian optimization (BO) issue, wherein it is required to efficiently trade-off between exploration and exploitation in order to optimize an unknown objective function.
The SAMPLE Platform
Self-driving Autonomous Machines for Protein Landscape Exploration (SAMPLE) is a technology that allows the quick engineering of proteins without subjectivity, feedback, or human interaction. SAMPLE is powered by an intelligent agent that creates new proteins to test theories and learns the links between protein sequence and function from data. Using a completely automated robotic system to synthesize genes, express proteins, and detect enzyme activity biochemically, the agent evaluates the designed proteins experimentally in the physical environment. Fully autonomous design-test-learn cycles are made possible by the intelligent agent’s seamless connection with experimental automation, which allows it to comprehend and optimize the sequence-function landscape.
Artificial Intelligence for Protein Engineering
The researchers created a fully autonomous system that would resemble the biological discovery and design process of humans. It is possible to think of human researchers as intelligent beings who behave in a lab setting and get data back as feedback. Human agents learn behaviors to accomplish engineering goals and gain a system understanding through frequent interactions with the laboratory environment. SAMPLE is an intelligent agent that explores protein sequence–function correlations and engineers proteins in a lab setting by learning on its own, making decisions, and acting.
The creation of proteins using a Gaussian process (GP) model is the intricate process known as protein engineering. A multi-output GP is used by the intelligent agent SAMPLE to model the properties of protein sequences. This model predicts thermostability for active sequences and has demonstrated good predictive capacity, with an 83% accuracy rate in the active/inactive classification.
In protein engineering, sequential decision-making under uncertainty is facilitated by the upper confidence bound (UCB) algorithm. Two BO heuristic techniques were developed to direct sampling in the direction of functional sequences. The Expected UCB technique uses the expected value of the UCB score to select the sequence with the highest expected UCB value, whereas the UCB positive method chooses sequences that the GP classifier predicts to be active.
The agent creates proteins and submits them for experimental evaluation to the SAMPLE lab setting. For automated gene assembly, cell-free protein expression, and biochemical analysis, a highly efficient and reliable pipeline is created. This process assembles DNA fragments that have already been synthesized, amplifies the result using polymerase chain reaction (PCR), and then determines which protein has the highest UCB value.
Conclusion
The SAMPLE platform is an automated laboratory that integrates automated learning, decision making, protein design, and testing to revolutionize synthetic biology and protein technologies. The platform is suitable for a wide range of protein engineering targets and functions, including enzymatic activity, thermostability engineering, selectivity, and novel chemical reactions. SAMPLE uses an unbiased approach to investigate the effect of sequence variation on function without prior knowledge of protein structure or mechanism. The biochemical testing required is the biggest obstacle to creating functional samples of new proteins. Automated systems can incorporate sophisticated analytical tools such as nuclear magnetic resonance spectroscopy or liquid chromatography-mass spectrometry to increase the range of protein functions to be designed. Thanks to its deployment in Strateos Cloud Labs, other synthetic biology researchers can now access and use SAMPLE’s experimental pipeline at a lower cost. With its agents exploring different regions and specializing in certain tasks, SAMPLE has the ability to accelerate and streamline the protein engineering process. It also shows promise for coordinating many agents towards a single protein engineering aim. Multi-agent coordination systems benefit further from the decentralized and on-demand nature of cloud laboratory environments.
Almost every industry is being disrupted by the potent combination of automation and artificial intelligence, from waste management, manufacturing, and food preparation to pharmaceutical research and agriculture. By automating extremely arduous, time-consuming, and wasteful protein engineering campaigns, self-driving labs will transform the areas of synthetic biology and biomolecular engineering. This will enable quick turnaround times and free up researchers to concentrate on significant downstream applications. As deep learning, robotic automation, and high-throughput instrumentation continue to progress, intelligent autonomous systems for scientific discovery will become more and more potent.
Article Source: Reference Paper
Follow Us
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.