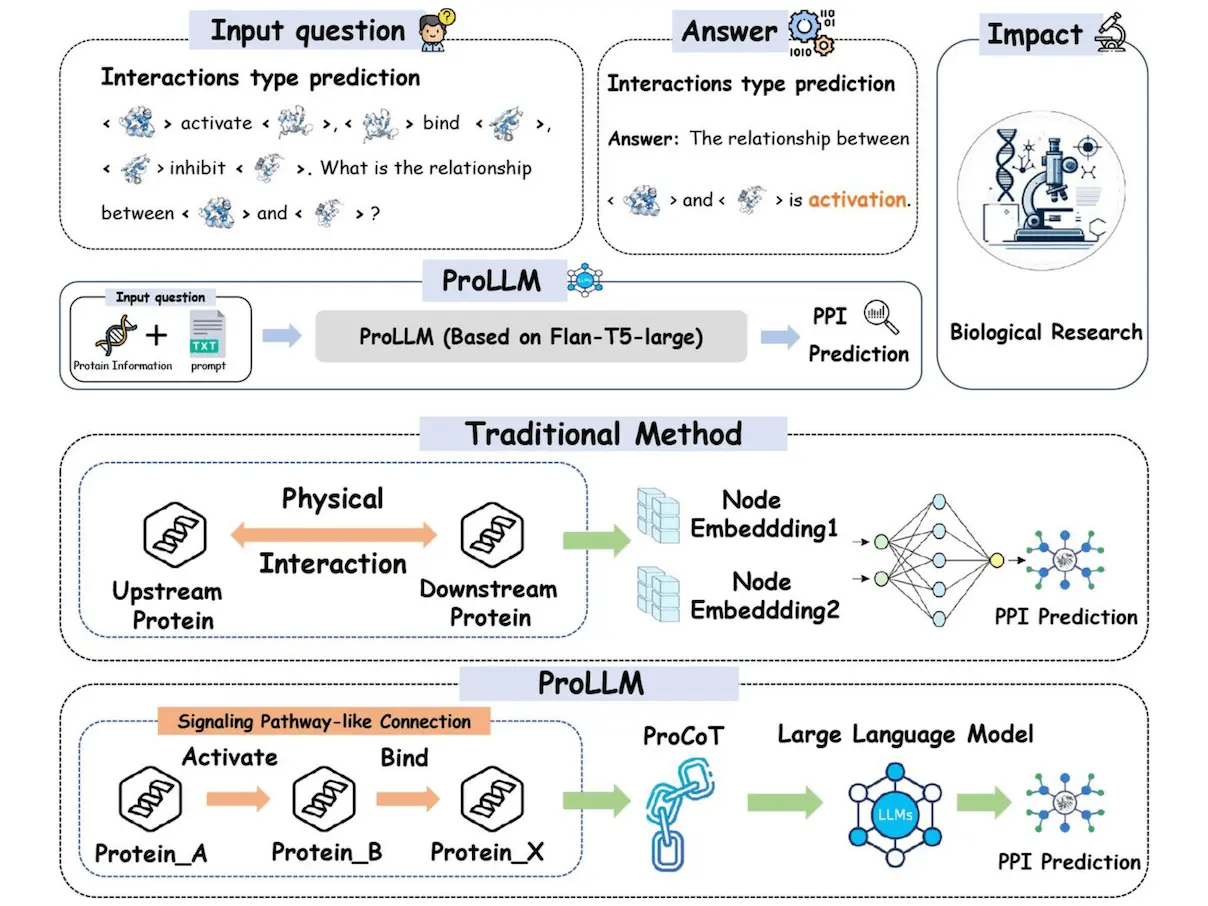
Predicting protein-protein interactions (PPIs) is essential to comprehend biological processes and illnesses. Conventional machine learning techniques ignore nonphysical linkages made through intermediary proteins in favor of direct physical interactions. By converting structured data into natural language prompts, mapping protein linkages into sentences, finding indirect connections, and following the journey from upstream to downstream, Large Language Models (LLMs) present a novel potential. As a result, the researchers present a brand-new framework called ProLLM that makes use of an LLM specifically designed for PPI. Researchers specifically suggest the Protein Chain of Thought (ProCoT), miming signaling pathways’ biological mechanism as natural language prompts. A protein reasoning method called ProCoT forecasts how upstream and downstream proteins will interact in biological signaling circuits. It trains ProLLM in the ProCoT format, which improves its comprehension of intricate biological issues. ProLLM also helps with instruction fine-tuning in protein knowledge databases and embedding replacement of protein locations in natural language prompts. Thorough validation against benchmark datasets demonstrates its effectiveness, demonstrating a notable improvement in prediction accuracy and generalizability over current approaches. This demonstrates how LLMs can change PPI and be an effective tool for a range of biological and medical research applications.
Introduction
When it comes to biological processes, including pharmaceutical research, protein-protein interactions (PPIs) are essential. PPI detection is accomplished by experimental techniques such as quantitative proteomics and yeast two-hybrid. Nevertheless, the labor- and time-intensive nature of wet-lab approaches for PPI prediction calls for more accurate computational techniques. Protein interaction research has witnessed the rapid growth of techniques such as Convolutional Neural Network (CNN) and Graph Neural Network (GNN) in computational biology in recent years. Protein amino acid sequences are converted into numerical vector representations via CNN-based methods like TAG-PPI, which then extract features for PPI tasks.
Unfortunately, the accuracy and interpretability of CNN approaches are limited by fixed receptive fields and poorly defined spatial correlations in protein sequences. Proteins are treated as nodes, and their interactions are seen as edges in GNN-based approaches such as GNN-PPI, which create a network made up of proteins. Although GNNs can use network structural information well, their development for protein prediction is limited because they are not as good at learning protein chains as transformer-based models are. They cannot fully capture and comprehend the relationships and dynamic changes in the signal-passing process in real organisms.
Large Language Models, including ProBert and ProteinLM, have also been employed in this PPI area in addition to the GNN and CNN techniques. Researchers can use the direct cosine similarity of the representation or train an MLP to make a PPI prediction, provided that these models can derive a protein representation. These techniques, however, still need to capture the protein chain interactions, such as signaling pathways, fully. Furthermore, prior research has solely employed LLMs as a feature extractor. Lately, it has been demonstrated that employing LLM as a link predictor outperforms conventional GNN baselines and is more effective at capturing relational information between nodes in knowledge graph tasks.
Understanding ProLLM and ProCoT
The drawbacks of current techniques that concentrate on single protein-protein interactions are addressed by ProLLM, a revolutionary method for predicting protein interactions in signaling networks. ProCoT is designed to directly anticipate the kind of protein interaction using a large language model to learn the rule of signal pathways. This method considers the cumulative impact of several protein interactions to address the ignorance of global, non-physical linkages between proteins.
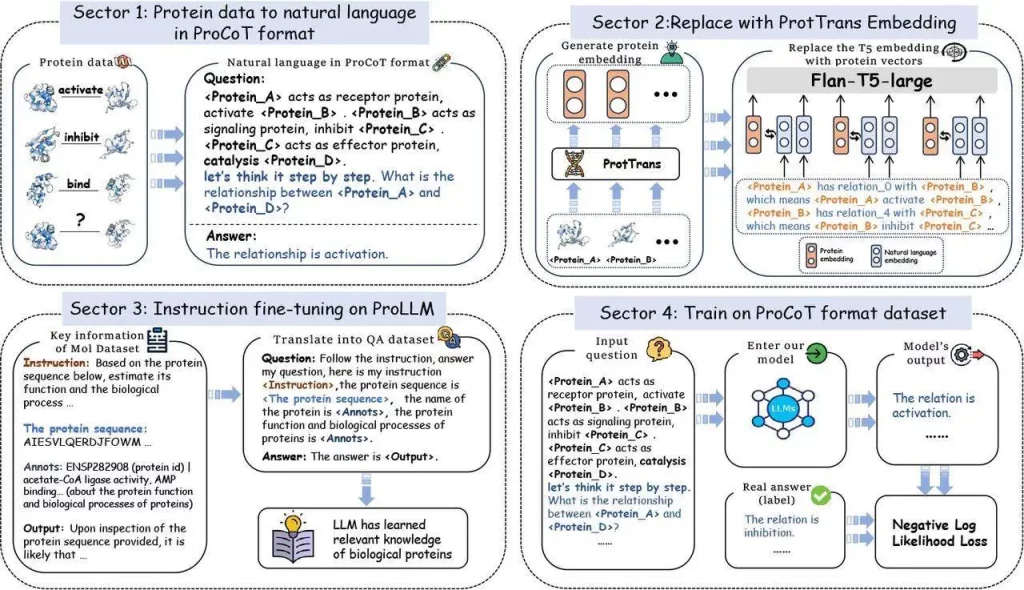
ProCoT is a data format that uses a thought-chain approach to model the signal transduction process, making it possible to forecast protein interactions in signaling pathway issues. It can articulate thought processes step-by-step, creating a chain of reasoning, and it takes this idea a step further by simulating protein signaling cascades in the domain of proteins, giving LLMs a richer understanding of proteins. By substituting embedding enriched with protein information for the conventional language model embedding, the method also tackles the problem of low protein comprehension in LLMs. This allows LLMs to reason about the direct interactions between proteins, answering queries concerning protein relationships—a crucial aspect of biological research—by infusing them with knowledge of the protein domain.
Understanding PPI and PPI Models
Protein-protein interactions (PPIs) are essential to all living things, and mass spectrometry, quantitative proteomics, synthetic lethality, and yeast two-hybrid experiments have all been used extensively in the study of PPI detection. As for Y2H experiments, they shed light on the roles and controls of proteins, and synthetic lethality pinpoints crucial gene pair interactions. Though it takes a lot of work, time, and resources, quantitative proteomics—especially with mass spectrometry—shows how dynamic the interactome is under different circumstances.
PT-GNN learns node properties from sequence and structure by integrating various data sources for link prediction. DeepRank-GNN provides a modular framework for predicting GNN-based interaction patterns. HIGH-PPI presents a hierarchical graph learning model for efficient molecular detail extrapolation and PPI prediction. The accuracy of biomolecule prediction is improved by geometric GNNs’ superior modeling of spatial nuances.
To predict how mutations may affect protein interactions, mmCSM-PPI collects detailed features. To improve the prediction accuracy of PPI, MAPE-PPI describes the microenvironment of amino acid residues in proteins and encodes it into discrete codes. The advancement of PPI predictions through machine learning techniques has greatly enhanced our comprehension of biological systems.
Protein sequence prediction has greatly improved with recent natural language processing (NLP) developments. Extensive textual datasets have been used to train large language models such as GPT, LLaMA, and T5, improving their performance on various NLP applications. Pre-trained on large-scale protein sequences, Protein Large Language Models (PLMs) like ESM, ProtTrans, and ProteinBert have revolutionized performance on a range of downstream tasks like N-linked glycosylation site prediction, single peptide prediction, structure prediction, and subcellular localization prediction. These models benefit bioinformatics, complicated network research, and generative link prediction applications.
The Reasons Behind ProCot’s Results: The Biological Instinct of ProCot
The AI-based system ProCoT is intended to mimic biological signaling pathways, which are a sequence of regulated, rule-based interactions that constitute a highly well-organized chain of information transfer. When managing serialized data, this pattern aligns with natural language processing (NLP) techniques. To better comprehend biological signal transduction processes, the system uses Flan-T5’s self-attention blocks to capture indirect interactions between proteins. The system also includes a signal interruption mechanism that imitates protein adaptation to ensure precise cell response to outside changes. ProCoT design is based on biological principles overall, which improves our understanding of biological signaling systems.
Conclusion
ProLLM is a unique framework that represents protein data in natural language forms and leverages Natural Language Models (LLMs) to predict protein-protein interactions. ProCoT has made significant contributions. Its sequential format, which LLMs are particularly good at understanding, translates complex protein signaling pathways into natural language prompts. ProLLM refines training on protein knowledge datasets and incorporates ProtTrans’s protein-specific embeddings. ProLLM surpassed existing graph-based and language model techniques in terms of prediction accuracy and generalizability, according to experiments conducted on four PPI datasets.
Article Source: Reference Paper | ProLLM code is available on GitHub.
Important Note: bioRiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.