A new NLP model called BioMedLM from the researchers of Stanford University and DataBricks seeks to rival models like Med-PaLM 2, which performs admirably on a range of biological tasks but is computationally costly, requires input data from the internet and is trained on unidentified data sources. It is possible to create smaller, more focused models by developing and publishing BioMedLM. Trained on PubMed abstracts and full articles, BioMedLM is a 2.7 billion parameter autoregressive model in the GPT approach that can outperform larger models in multiple-choice biomedical question answering. It received a score of 69.0% on the MMLU Medical Genetics test and 57.3% on the MedMCQA (dev). When BioMedLM is optimized, it can offer clear, private-protecting, affordable, and eco-friendly bases for NLP uses in biomedicine.
Introduction
The natural language processing sector is changing because of large language models like OpenAI’s GPT-4. These models are quite good at summarising and answering questions because they were trained on big corpora and improved with human feedback. Because they make it possible to retrieve and summarise data from clinical records, radiology reports, and biomedical literature, they may have an impact on biomedical research and healthcare. Domain-specific language models are a prominent technology in the medical profession because they can improve patient care, lower healthcare costs, and speed up biomedical discoveries.
Drawbacks of Large Language Models
- Access to these tools may be challenging for organizations due to rising compute and inference expenses since 2015 and API fees.
- These models must be run on massive computing clusters and accessible remotely via the internet due to their enormity and corporate secrecy. Sensitive data must be transmitted in order to remove the chance of on-device inference. The requirement to use corporate APIs to access these models presents concerns over the disclosure of personally identifiable information (PII) to third parties. This runs counter to the Health Insurance Portability and Accountability Act’s (HIPAA) requirements for patient data privacy in the medical field.
- These models’ restricted nature also leads to a lot of issues. Any company that uses these concepts is ultimately dependent on a large technology company. Any services an organization develops on top of the model may be instantly terminated if the tech company stops supporting it for any reason.
- The fact that the training data for business models is a closely kept secret is one of the most notable disadvantages of this closed nature. This puts a company that uses these models at risk from a business standpoint. For instance, an AI model provider’s model can suddenly stop working if it is trained on copyright-violating data. From the standpoint of a practitioner, the quality and dependability of model replies are further clouded by the lack of openness surrounding model training data. Furthermore, from the standpoint of research, it is difficult to examine the connection between training data and task performance downstream when one is unaware of the makeup of the model training data.
Introducing BioMedLM
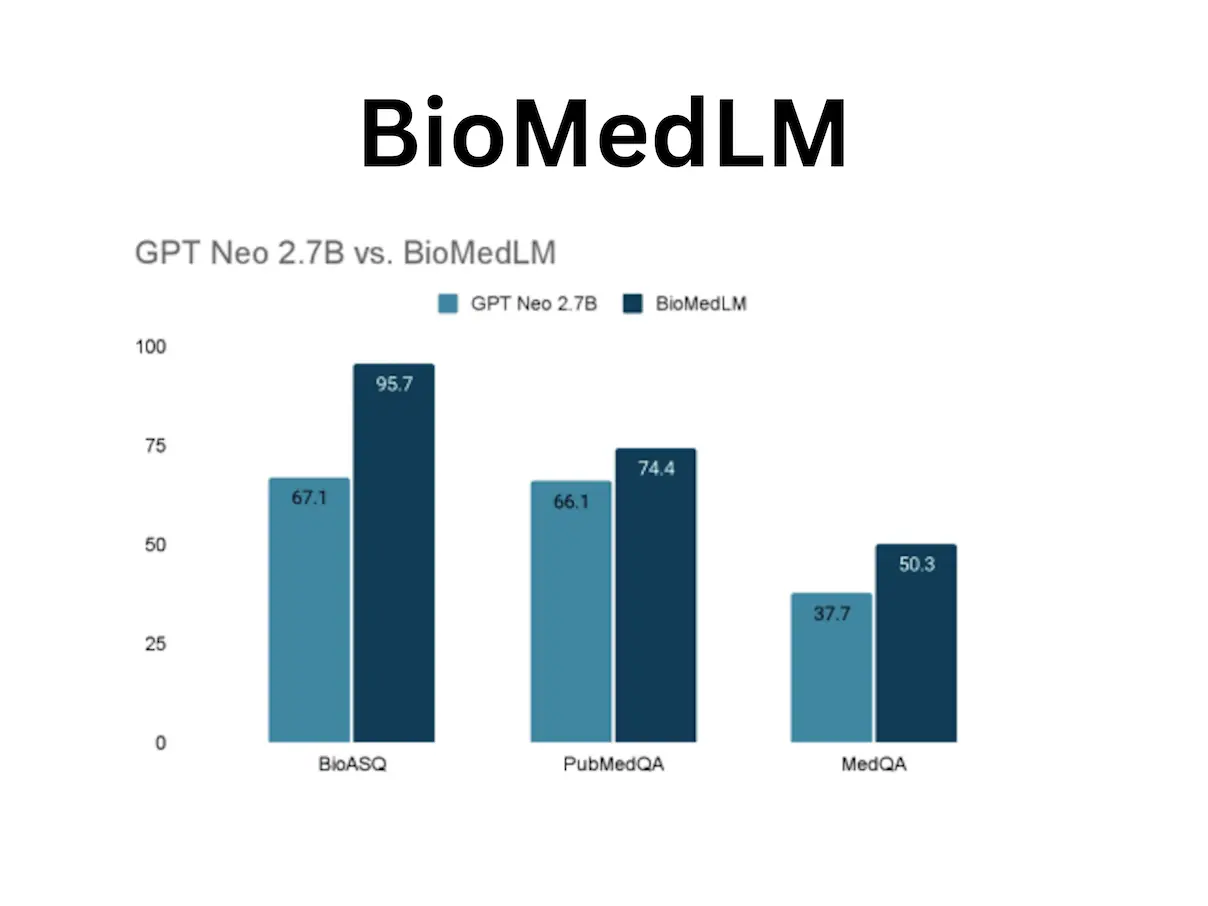
A promising path in the field of biomedical text analysis is represented by BioMedLM, a 2.7 billion-parameter biomedical language model. Designed only for PubMed abstracts and entire articles, it provides a more condensed approach to biomedical text training by concentrating on domain-specific performance. This model, an autoregressive model in the GPT-2 type, presents a viable way to improve the caliber of biomedical text analysis. The MMLU Medical Genetics model BioMedLM has shown promising results in answering multiple-choice biomedical questions, outperforming larger models such as GPT-Neo 2.7B. The BioASQ, PubMedQA, and MedQA tests show increased accuracy for the model.
Additionally, it can generate multi-sentence responses from the HealthSearchQA dataset on medical-related topics. Because BioMedLM is tiny, it can execute inference on a laptop and be fine-tuned on a single A100 GPU, allowing organizations to serve it internally without transferring sensitive data outside their networks. Its open nature allows anybody to obtain and adjust the training data as they see fit, and its fully documented training data offers insights into the model’s performance.
Businesses are keeping the specifics of their language model architectures and training under wraps as the AI race heats up. In addition to adding to the corpus of information regarding the training of domain-specific models and the application of language models to biomedical NLP tasks, BioMedLM seeks to disseminate knowledge about the construction of language models. BioMedLM offers a helpful tool for language analysis and production in biomedical jobs by offering a smaller, more performant model.
Training of model BioMedLM
BioMedLM was trained using 34.6 billion tokens from the Pile dataset. The training run used MosaicML Cloud for heavy workloads, completing 8.67 runs across the data. The Composer training library, PyTorch FSDP, Hugging Face’s GPT-2 model code, and Flash Attention, a memory-reducing method, were used to pre-train the model. In a total of 6.25 days, the training run was finished. The model was trained using Decoupled AdamW with batch size 1024 and sequence length 1024 for 300 billion tokens to minimize cross-entropy loss. Training loss divergences were consistently observed in the first trial runs at the 1.5 billion scale; however, these were eliminated when bf16 was used. The final training process of the 2.7 billion parameter model did not suffer from any divergences. Despite having more passes through the data than comparable models, the model showed steady perplexity improvements and improved downstream task performance. The study concludes that training for the full 300B tokens was worth it for language model perplexity and downstream task performance.
BioMedLM Application
BioMedLM, launched in December 2022, has demonstrated accuracy of 50.3% and 74.4% on a variety of biological benchmarks, including medical quality assurance (QA) activities (MedQA and PubMedQA). It has been applied to enhance the transferability of models for classifying clinical note sections, including progress notes, discharge summaries, and colorectal clinical notes. Additionally, BioMedLM has demonstrated state-of-the-art performance in named entity recognition for broad medical applications utilizing the bio-entity tasks JNLPBA and NCBIDisease.
Additionally, relation extraction and categorization using BioMedLM have been tested in a number of biomedical fields, such as protein interactions and pathways. BioMedLM performed the best in STRING Task 2, outperforming larger models such as LLaMA, Alpaca, and BioGPTLarge. BioMedLM was optimized to extract microbiome-disease interactions in the microbiome domain, outperforming GPT-3 (0.810). In genetics, BioMedLM has been evaluated on the GeneTuring test, focusing on nomenclature, genomic location, functional analysis, and sequence alignment.
In the multi-modal space, BioMedLM has been used in vision-language pre-training models for computer-aided diagnosis and prefix tuning for visual question-answering tasks.
Conclusion
The low-cost model BioMedLM performs exceptionally well in biology-related tasks because it was trained using PubMed domain knowledge. This information improves accuracy and decreases false positive rates in extraction tasks, as well as performance in tasks involving medical text corpora. The model is a useful tool for a variety of activities because of its medium size and emphasis on domain knowledge.
Article source: Reference Paper | BioMedLM model is available on the Hugging Face | GitHub
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.