Drug discovery is one of the many fields where recent developments in conversational large language models (LLMs), like ChatGPT, have shown incredible promise. However, the majority of the work that has already been done is on examining conversational LLMs’ skills with regard to chemical reactions and retrosynthesis. However, there is still much to learn about drug editing, a crucial step in the drug development process. In order to fill this gap, the researchers from Mila-Quebec Artificial Intelligence Institute present ChatDrug, a framework that makes it easier to conduct a thorough inquiry into drug modification utilizing LLMs. To facilitate efficient drug editing, ChatDrug combines three modules: a prompt module, a retrieval and domain feedback (ReDF) module, and a chat module. The empirical findings demonstrate that ChatDrug achieves optimal performance on 33 of 39 drug editing tasks, including peptides, proteins, and small molecules.
Introduction
Artificial intelligence (AI) tools have revolutionized drug discovery in recent years. They offer enormous potential for speeding and improving different stages of the process, such as virtual screening, lead optimization, reaction and retrosynthesis, protein folding, and inverse folding.
However, a large portion of the research that has already been done has just looked at drug structure data, treating the medications’ fundamental chemical structure as a single modality. The drug discovery pipeline, on the other hand, uses iterative refining procedures that involve speaking with subject matter experts to incorporate their input and eventually provide the desired result. However, remarkable progress has been achieved in large language models, which have remarkable capacities for comprehending human knowledge and potential thinking abilities. These findings motivate people to look into the possibility of using LLMs’ reasoning and conversation skills for multimodality AI-assisted drug discovery.
Understanding Drugs
Substances that are used to treat, diagnose, prevent, or alleviate the symptoms of an illness or other abnormal state are referred to as drugs. The three most widely used medications—small molecules, proteins, and peptides—were examined in this study. Groups of atoms joined by covalent bonds make up small molecules. Molecular graphs and SMILES (simplified molecular-input line-entry system) strings are examples of frequently used data structures. In this paper, researchers employed the SMILES strings in ChatDrug. Twenty amino acids, each of which is a tiny molecule, make up proteins, which are complicated macromolecules. Peptides are a unique class of protein made up of short chains of amino acids. Researchers focused on the drug editing task in this work. Drug editing is the lead optimization or protein design step in the process of drug discovery.
ChatGPT in Drug Editing
Drug development could benefit greatly from conversational learning models (LLMs) because of their extensive knowledge base, quick adaptation, and interactive communication. These models, such as ChatGPT, are used in a number of industries, including drug development. They can provide more accurate responses by rapidly activating pertinent pre-training concepts. Furthermore, a dynamic flow of information is made possible by its interactive communication function, which incorporates comments from domain experts or past knowledge. In drug discovery tasks, this reciprocal flow of information enhances relevance and accuracy. Drug editing, which entails changing drug substructures, is one difficult endeavor in the drug development process. Conventional approaches that depend on subject matter specialists may be prejudiced or opinionated. While recent research investigates text-guided drug editing in other modalities, they do not have the conversational potential of ChatGPT.
Diving into ChatDrug
ChatDrug is a platform designed to improve drug editing and open new options using contrastive LLMs like ChatGPT. Users can initiate a conversation with LLMs, including domain knowledge, and insert such retrieved information into the dialogue within the ChatDrug framework for drug editing. To enable robust prompt engineering from LLMs, ChatDrug first implements a PDDS (prompt design for domain-specific) module. ChatDrug incorporates a module for retrieval and domain feedback known as ReDF. Through the utilization of the extensive domain knowledge at hand, a ReDF module of this kind facilitates timely updates and enhances the model’s ability to produce correct results. ChatDrug takes a conversation-based strategy that fits with the pipeline’s iterative refining approach to drug discovery. A dynamic and cooperative process is made possible by such an interactive schema, which successfully incorporates domain experts’ feedback to produce the intended results.
ChatDrug is a distinct drug editing tool that has two main features: a compositional feature that breaks down difficult concepts into smaller ones and an open vocabulary feature that lets users explore novel drug concepts outside of a predefined set of annotations. This makes tackling difficult drug editing jobs easier. A benchmark covering a wide range of tasks that can be computationally analyzed and involves indeterministic answers is required to confirm ChatDrug’s efficacy. Over the course of three popular drugs, the tool presents 39 editing tasks: 28 for small molecules, 9 for peptides, and 2 for proteins.
ChatDrug Framework
- Prompt Design for DomainSpecific (PDDS) Module – ChatDrug is a novel drug editing method that focuses on generalizing a natural language model (LLM) trained on certain types and sources of data. When altering medications, this technique is particularly effective with small molecules, proteins, and peptides that attach to proteins. Text prompts from ChatDrug are designed to enable domain-specific actions with reasonable processing costs. Drug editing goals should focus on high-level qualities rather than precisely altering substructures, as this approach is better suited for properties-related and fuzzy matching concerns. The rapid design takes into consideration properties like peptide-MHC binding for peptides, secondary structure for proteins, drug-likeness, permeability, and the number of donors/acceptors for tiny molecules.
- Retrieval and Domain Feedback (ReDF) Module – The ReDF module is a crucial component that utilizes retrieval databases and domain knowledge. It incorporates conversational LLMs’ language comprehension skills by extracting relevant material from the database and putting it into the text prompt. For each input medication and prompt, a candidate drug x˴ is generated based on the issue design. ReDF produces a drug xR that fulfills the domain and similarity feedback functions. This injection, which is similar to the in-context learning (ICL) paradigm, improves performance by mapping ground truth data-label pairs and illuminating the in-distribution data and label space.
- Conversation module – The interactive nature of conversational LLMs, such as ChatGPT, is a compelling feature. This allows the LLMs to add previous knowledge and update the results iteratively.
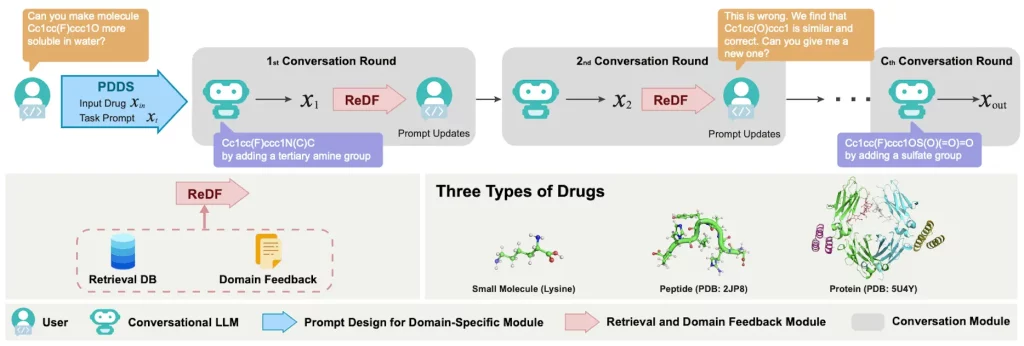
Image Source: https://doi.org/10.48550/arXiv.2305.18090
Conclusion
ChatDrug is a framework that utilizes ChatGPT for drug editing tasks, aiming to alleviate knowledge redundancy among knowledge. It can extract domain-specific information for editing molecules and summarize them into five rules. However, ChatDrug faces limitations in understanding complex drug structures, such as 3D geometries, which may require more geometric modeling. Additionally, it requires certain conversational rounds to reach strong performance. To address these limitations, ChatDrug can play a positive role in knowledge summarization, aiming to reduce computational costs using ChatGPT’s knowledge summarization ability. According to this research, ChatDrug is capable of identifying crucial substructures for drug editing, including protein structures, peptide patterns, and functional groups in molecules. This study highlights the potential of conversational LLMs and ChatGPT for drug editing, improving interpretability, and enabling well-informed decision-making. This study opens the door for a more effective and cooperative drug discovery pipeline, which advances pharmaceutical research and development. Overall, ChatDrug is a promising direction for machine learning and drug discovery communities in drug editing tasks.
Article Source: Reference Paper | ChatDrug code is available on GitHub | Website
Follow Us
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.
















