The class B scavenger receptor (SR-B) proteins, which are essential for innate immunity, pathogen identification, apoptotic cell clearance, and metabolic sensing, have an ectodomain called the CD36 domain. Previous research has suggested that SR-B proteins are old, although their diversity within Eukarya is yet unknown. The University of Miami researchers examined SR-B homologs found in the transcriptomes and genomes of 165 different eukaryotic species. The existence of highly conserved amino acid motifs across key eukaryotic supergroups supports the presence of an SR-B homolog in the last common ancestor of eukaryotes. Within the CD36 ectodomain, a conventional asymmetric beta barrel tertiary structure is retained across Eukarya, according to comparative investigations of SR-B protein structure. The development of SR-B ligand-sensing specialization across several eukaryotic clades may be reflected in the multiple occurrences of lineage-specific sequence expansions in the CD36 apex region.
Introduction
Cellular reactions are frequently triggered by external stimuli via membrane-bound pattern recognition receptors (PRRs). The “CD36 family,” or SR-B proteins, are a functionally defined class of transmembrane receptor proteins that can function as PRRs and recognize a broad range of ligands. Two transmembrane domains involved in intracellular signal transduction and an ectodomain domain that makes up the CD36 antigen distinguish these proteins from one another. The SR-B proteins exhibit expression in a range of cell types and have the ability to bind a wide variety of ligands, including long-chain fatty acids, MAMPs, lipid-based pheromones, vitamins, cell adhesion proteins, cholesterols, and phospholipids. The abnormalities associated with Alzheimer’s disease in mammalian microglial cells may be related to the dysregulation of CD36.
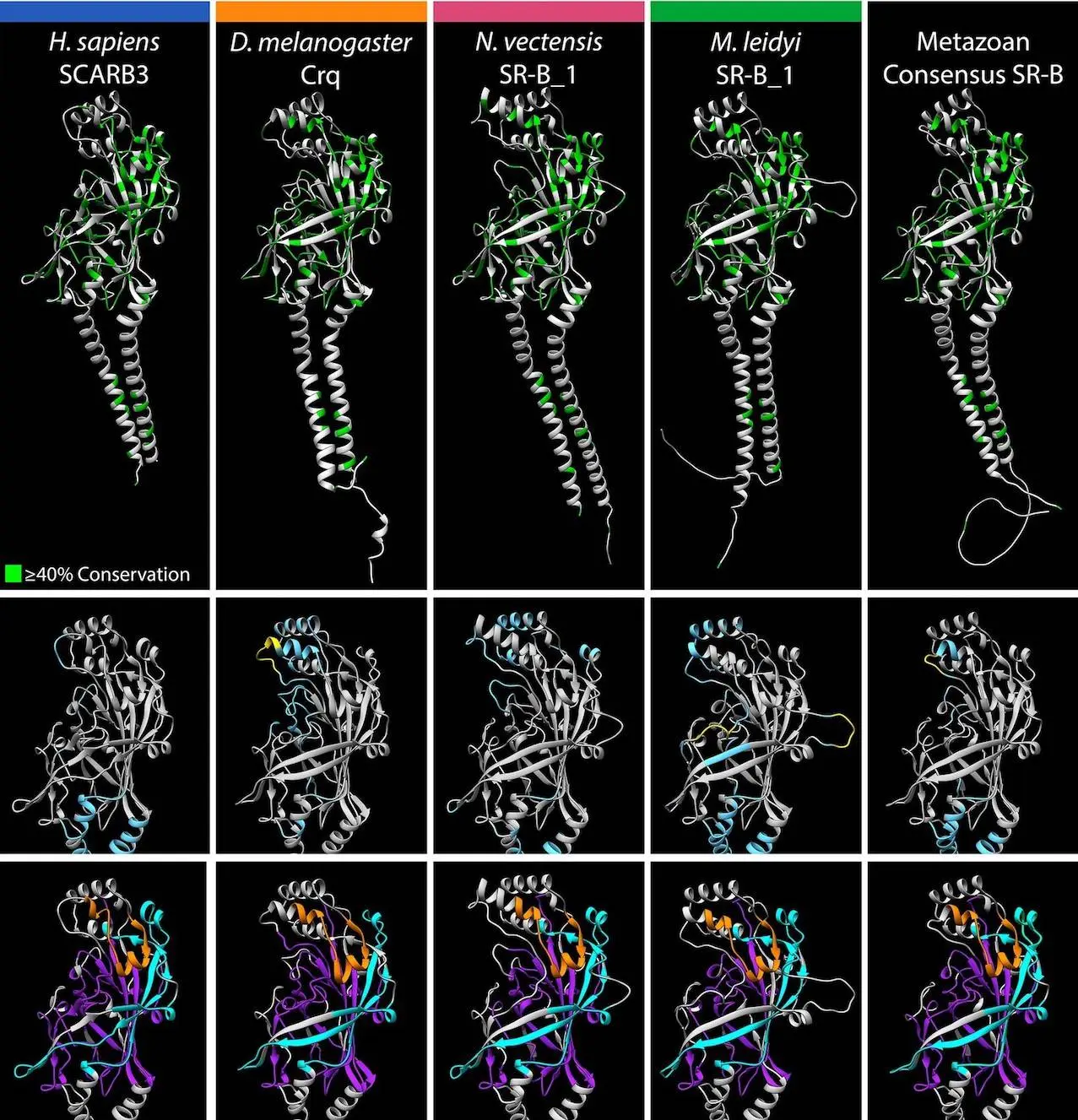
The human SR-B proteins’ X-ray crystallography shows that the CD36 ectodomain folds into an asymmetric beta-barrel structure with a cluster of alpha helices at the apex. This “three-helix bundle” is essential in recognizing ligands. Transport of LCFAs and cholesterol esters is facilitated by the ectodomain’s proximity to a binding pocket and intramolecular tunnel. Many metazoans, such as mammals and sponges, have had the functional characteristics of their SR-B proteins studied. Three different lineage-specific clusters of proteins containing the CD36 domain that share structural similarities are found in insects.
The study analyses SR-Bs from different eukaryotic lineages using sequence alignments, motif prediction, and artificial intelligence. The last eukaryote common ancestor (LECA) has an ancestral SR-B-like protein, according to the results. SR-Bs have a similar beta-barrel tertiary structure in the CD36 ectodomain, but the membrane-distal apex region has an evolutionarily labile area. Aa residues essential for ligand recognition are present in the apex region, indicating that sequence expansion and structural divergence may be reflective of the evolution of SR-B ligand selectivity.
Understanding CD36 domain
Class B scavenger receptors (SR-Bs), also called the “CD36 family,” are distinguished from other SRs by having an ectodomain domain that contains the CD36 antigen and two transmembrane domains with relatively short N- and C-terminal cytoplasmic tails that are involved in intracellular signal transduction. Several multifunctional SR-Bs have been identified in mammals, including CD36 (sometimes referred to as SCARB3), lysosomal integral membrane protein type 2 (LIMP-2 or SCARB2), and SCARB1. Vertebrate SR-B proteins are expressed in a range of cell types and bind a wide range of ligands, such as long-chain fatty acids (LCFAs), cholesterol, phospholipids, lipid-based pheromones, lipid-soluble vitamins, cell adhesion proteins, microbe-associated molecular patterns (MAMPs), and damage-associated molecular patterns (DAMPs) from cell debris.
Alzheimer’s disease pathology may entail the deregulation of CD36 in mammalian microglial cells. Along with working with other cell surface PRRs, SR-B proteins that are capable of recognizing MAMPs and DAMPs also facilitate innate immune responses, such as the creation of phagosomes and endosomes.
Presence of SR-B-Like Proteins across Diverse Eukaryotes
A CD36 ectodomain surrounded by two transmembrane helices characterizes SR-B protein sequences operationally. In 165 publically accessible eukaryotic proteomes, the study employed HMMER to look for CD36 domains. It found 418 distinct protein sequences that featured CD36 domains. True SR-B homologs numbering 279 were found in these sequences. Eukaryotic lineages that included Hemimastigophora, Ancyromonadida, and CRuMs were among those from which SR-B homologs were found. Amorphea contains SR-B homologs in all major clades, including Amoebozoa, Holomycota, Ichthyosporea, Metazoa, and Choanoflagellata. Apusomonadidan Thecamonas trahens, pluriformean Corallochytrium limacisporum, Breviatea, and Tunicaraptor unikontum all had transcriptomes that were shown to have SR-B homologs.
Although no SR-B homologs were found in streptophytes, glaucophytes, or rhodophytes, it appears that each of these lineages experienced distinct losses. SR-B homologs were found in seven species within Archaeplastida that are related to chlorophytes. Based on the study, SR-B homologs were found in Stramenopila, Alveolata, and Rhizaria, the three primary clades that make up the SAR supergroup.
Protein domains not generally associated with SR-Bs were present in certain recovered SR-B protein sequences, which included cytoplasmic portions that were abnormally lengthy. These sequences seemed to reflect gene model fusions. Moreover, CD36 ectodomain aa sequences were extracted and aligned from bona fide SR-B homologs that were found. This analysis demonstrated a notable variance in sequence length between eukaryotic CD36 ectodomains based on lineage.
Evolutionary Insights from Phylogenetic Analyses
The links between the CD36 domains of SR-B homologs across Eukarya were examined using maximum likelihood and Bayesian phylogenetic analysis. Shallow nodes that link SR-B orthologs and indicate lineage-specific paralogs, including several informative clades across metazoans, were shown to have congruent bootstrap and Bayesian posterior probability (BPP) support.
Nonetheless, it is still unclear how other metazoan SR-Bs relate to emp-like, crq-like, and SNMP-like insect SR-Bs. Only the oomycetes in the Peronosporales and Pythiales orders include the biggest supported clade of nonmetazoan SR-Bs. SR-Bs grouped into two well-supported clades among amoebozoans, one with putative homologs of LmpB and the other with homologs of LmpA and LmpC. The combined information indicates that eukaryotic lineages have undergone significant sequence divergence in SR-Bs, which has obscured homologous links between lineages during their evolution and diversification. Despite taxonomic sampling bias, the phylogeny reconstruction supports the presence of at least one ancestral SR-B in LECA.
Diversification of the CD36 Domain “Apex”
Significant structural heterogeneity among eukaryotic lineages associated with the CD36 apex is shown by the study. SR-B homologs with CD36 ectodomains longer than 500 aa show distinct apex regions and sequence expansions peculiar to each lineage. These sequence expansions coincide spatially with helices α4 and α5 of the ligand-interacting three α-helix bundles in human SCARB3, and they occur within the same relative location between structural features associated with motifs M4 and M6.
Sequence expansions are also seen in some lineages close to motif M6, which corresponds geographically with α7, the third helix in the human SCARB3 three-helix bundle. Despite relatively low sequence conservation, the apex region of SR-Bs with CD36 ectodomains <500 aa is characterized by a comparable tertiary structure. Structural comparisons reveal some variation in α-helix length and orientation, with instances of significant primary sequence divergence despite tertiary structure conservation associated with presumed ligand-interacting features of the CD36 ectodomain.
Conclusion
Innate immunity, reproduction, and nutrition uptake are just a few of the biological activities that depend heavily on ligand interactions. Cell responses are triggered by integral receptor proteins, which identify external signals. The membrane-distal apex region of the CD36 domain may have undergone structural modification as a result of physiological and environmental ligand interactions. New tertiary structures have been produced as a result of sequence expansions in the ligand-sensing apex that are distinct to certain lineages. A more comprehensive understanding of this ancient receptor family’s evolutionary history can be obtained by comparative protein structure analysis.
Article Source: Reference Paper
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.