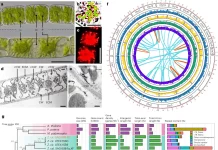
The dynamic localization of thousands of unique regulatory proteins to specific DNA sequences regulates the expression of genes. It has been a longstanding objective of molecular biology to comprehend this cell-type specific mechanism. However, it is still difficult because the majority of DNA-protein mapping techniques only look at one protein at a time. A study was conducted to overcome this problem by a group of researchers who introduced a split-pool-based technique called ChIP-DIP (ChIP Done In Parallel), which allows hundreds of different regulatory proteins to be simultaneously mapped throughout the genome in a single experiment. Researchers show that all classes of DNA-associated proteins, such as transcription factors, RNA polymerases, chromatin regulators, and histone modifications, produce extremely accurate maps when generated by ChIP-DIP. By analyzing these data, scientists can identify different types of regulatory elements and their functional activity by defining quantitative combinations of protein localization on genomic DNA.
Introduction
Thousands of regulatory proteins that localize at specific DNA sequences to activate, repress, and modulate transcription levels are involved in the regulation of gene expression specific to cell types. The Cell-type-specific chromatin states are defined by histone modifications and chromatin states, which are managed by chromatin regulators. These regulators regulate nucleosome location, DNA accessibility, and the reading, writing, and erasing of particular histone modifications. Molecular biology has spent decades trying to figure out how regulatory protein binding results in the expression of genes that are particular to a given cell type. The cell-type specificity of regulatory proteins and histone modifications’ interactions makes genome-wide mapping of these alterations difficult.
Early efforts to investigate cell type-specific regulation resulted in the creation of global consortiums such as ENCODE, PsychENCODE, and ImmGen. However, the majority of mammalian cell types, experimental and disease models, and model organisms remain uncharacterized because gene expression programs and protein binding maps are intrinsically cell type-specific. A highly scalable, multiplexed protein profiling technique is required to solve this, one that can profile a wide range of DNA-associated proteins, including difficult classes like transcription factors, and enhance protein mapping throughput by orders of magnitude. This would make it possible to map any sort of cell quickly and affordably, allowing for the investigation of important issues.
Understanding ChIP-DIP
An experiment can simultaneously map hundreds of different regulatory proteins thanks to the scalable platform known as chromatin immunoprecipitation done-in-parallel (ChIP-DIP). Data from over 180 DNA-associated proteins in human or mouse cells were generated by researchers using ChIP-DIP. Irrespective of the quantity of antibodies or protein composition, the platform produces precise genome-wide maps that are on par with conventional techniques and demonstrate great resilience. It has the ability to map every class of protein related to DNA, such as transcription factors, chromatin regulators, RNA polymerases, and histone modifications. For highly multiplexed mapping of protein localization within single cells, ChIP-DIP can be combined with an existing split-pool technique.
Applications of ChIP-DIP
- Mapping DNA-associated proteins
A technique for mapping hundreds of DNA-associated proteins in a single experiment is the ChIP-DIP experiment. Individual antibodies are coupled to beads using a distinct oligonucleotide tag, sets of various antibody-bead-oligo conjugates are combined, ChIP is carried out using this pool, split-and-pool barcoding is carried out, DNA is sequenced, and split-pool barcodes shared between genomic DNA and the antibody tag are computationally matched. A ChIP-DIP experiment yields data similar to that of a conventional ChIP-Seq experiment but with unique maps for each antibody used. Four well-studied proteins were used in the experiment: RNA Polymerase II enzyme, H3K4me3 histone modification, CTCF sequence-specific DNA binding protein, and H3K27me3 histone modification. The experiment was conducted on human K562 cells. The findings were associated with ChIP-Seq profiles produced by the ENCODE consortium and demonstrated consistent localization patterns at particular genomic locations. ChIP-DIP produces data similar to that produced by conventional technologies and is incredibly robust.
- Accurately maps hundreds of diverse DNA-associated proteins
The goal of the study was to investigate how well ChIP-DIP maps proteins from various categories, some of which have historically proven more difficult to map than others. In six trials, the researchers used ChIP-DIP to analyze more than 60 different proteins in human K562 cells and more than 160 different proteins in mouse embryonic stem cells (mESCs). The findings demonstrated that ChIP-DIP reliably maps histone modifications with unique patterns across the genome, signifying unique activity states and localizing at unique functional sites. A precise mapping of chromatin regulators (CRs) was also achieved; 67 CRs were linked to different signs of DNA methylation, acetylation, ubiquitination, and histone methylation. Additionally, the study discovered that ChIP-DIP can precisely map chromatin regulators from various complexes with unique functional characteristics.
ChIP-DIP also provided an accurate map of transcription factors (TFs), with 43 TFs in mESC and 15 TFs in K562. Accurately identifying DNA binding motifs for every TF, the researchers obtained high-resolution binding maps for TFs in both cell types.
Also, the three RNA polymerases and the post-translationally altered forms of RNAP II were successfully mapped by the researchers, proving that ChIP-DIP precisely maps the localization of RNA polymerases at different gene classes and characteristics. These findings validate ChIP-DIP as a highly-multiplexed, modular approach to producing high-quality maps for a broad class of DNA-associated proteins covering a variety of biological roles.
Conclusion
Using the molecular biology method known as ChIP-DIP, hundreds of regulatory proteins can be mapped to genomic DNA in a single experiment. This approach eliminates the need to choose between mapping a large number of marks in a small number of cell types or a small number of marks in a large number of cell types, enabling a thorough understanding of gene regulation and the consequences of genetic variations linked to human disease. By employing different sets of antibody-oligo-labeled beads to map protein binding inside numerous samples simultaneously, ChIP-DIP reduces the biological and technical variability involved with processing individual proteins and samples. Large-scale mapping of dynamic protein localization across several cell types and time points will be made possible by the capacity to assess regulatory proteins at scale, in a variety of cell circumstances, and with fewer sources of variability. To create additional capabilities, such as mapping 3D genome structures surrounding individual protein binding sites and enabling direct measurements of transcriptional activity and protein binding at individual genomic locations, ChIP-DIP can be directly integrated into a variety of existing split-pool approaches.
Article Source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.