Understanding biological processes and offering organized representations of intricate relationships are made possible by Knowledge Graphs, or KGs. However, the resources available today to mine KGs from biomedical literature are insufficient. Though they present a promising approach, large language models (LLMs) like GPT-4 have not yet been fully investigated in terms of their potential for end-to-end KG creation, especially for Molecular Regulatory Pathways (MRPs). The purpose of this work was to investigate the potential of GPT-4 for the comprehensive creation of a context-aware relational graph that can precisely identify context-specific MRPs of m6A methylation in a given sentence. ReguloGPT is a novel GPT-4 based in-context learning prompt that was created by the University of Pittsburgh researchers specifically for end-to-end joint name entity identification. The method addresses semantic discrepancies by embedding context within relational edges and encapsulates the hierarchical structure of MRPs. Using a benchmark dataset of 400 annotated PubMed titles on N6-methyladenosine (m6A) regulations, the study showed a significant improvement over previous techniques. Additionally, a unique G-Eval scheme utilizing GPT-4 for annotation-free performance evaluation was devised and shown useful in clarifying the regulatory processes of m6A in different types of cancer.
Introduction
Understanding molecular regulatory pathways—which show how chemical stimuli or genetic differences affect biological processes and diseases—is essential to comprehending biological functioning. By examining MRPs, researchers can pinpoint dysregulations that contribute to disease and direct the creation of tailored treatments. Knowledge Graphs (KGs) are currently essential for arranging and deciphering large amounts of data found in MRPs, including the relationships between genes, proteins, and biological processes. Databases such as KEGG, Reactome, and QIAGEN have been built through human curation; nonetheless, the number and pace of new research articles present a challenge. Biomedical knowledge may now be better extracted from literature using automated natural language processing (NLP) techniques, such as those found in INDRA, MSI, Hetionet, RepoDB, and DrugMechDB.
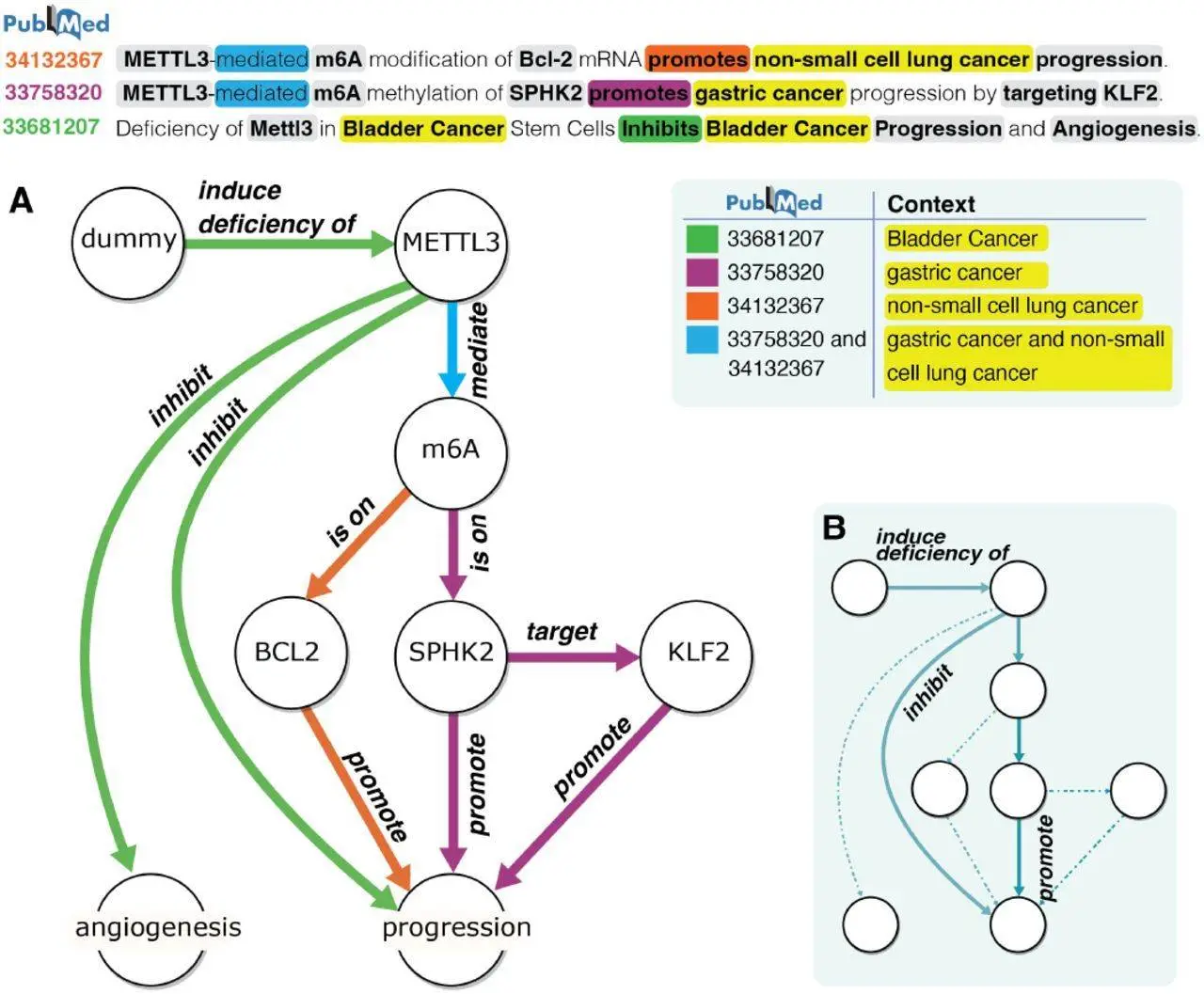
For the purpose of mapping complicated Multi-Relational Programmes with intricate linkages and hierarchical structures, current gene association mining techniques are insufficient. The statement “METTL3-mediated m6A methylation of SPHK2 promotes gastric cancer progression by targeting KLF2″ elucidates an N-ary relationship graph that is relevant to the context and involves multiple entities. Such intricate graphs are difficult to extract from MRP descriptions using current Natural Language Processing (NLP) techniques and need context identification, sophisticated Named Entity Recognition (NER), and N-ary Relationship Extraction (RE). Biomedical KG creation using existing NLP techniques can be divided into rule-based, machine-learning-based, or hybrid approaches like the Turku Event Extraction System (TEES). Nevertheless, these approaches struggle with N-ary interactions in MRPs and concentrate on binary relationships, which causes cascading errors and increases complexity.
EIDOS and TEES integrate rule-based techniques with machine learning, whereas REACH uses a rule-based methodology to detect items and relationships within biomedical texts. GNBR (Global Network of Biomedical Relationships) is an expert in using machine learning to extract gene-related information from biomedical literature efficiently. Its specialties include normalizing gene mentions and extracting binary relations. SemRep is an additional system that applies a rule-based approach to biological connection extraction, emphasizing meaningful semantic relation extraction. According to the study, METTL3 plays a role in myeloid leukemia, lung cancer, and breast cancer tumor metastasis. Based on data from five titles (PMID: 32766145, 36069931, 36609396, 34312368, 35319018), the sub-KGs unveil a complex dual-pathway mechanism. By specifically targeting METTL3, therapeutic intervention may be able to disrupt critical metabolic and structural pathways that are necessary for the spread of cancer. Tumor suppressor gene MEG3 is also detected in sub-KGs with myeloid leukemia and lung cancer.
Understanding reguloGPT
ReguloGPT utilizes PubMed phrases that illustrate biological regulatory processes to construct a context-aware knowledge graph (KG). The KG incorporates retrieved regulatory contexts and related PubMed IDs into edges, mirroring the hierarchy of biological pathways. This makes it possible to define context-specific regulation. In an effort to precisely comprehend context-specific MRPs that contain both explicit and implicit rules, researchers suggested reguloGPT, a GPT-4 driven ICL prompt, especially intended for end-to-end joint NER, N-ary RE, and context identification. The baseline, few-shot, and Chain-of-Thought (CoT) prompts for reguloGPT were created by the researchers. It is a unique method for extracting MRPs from literature using GPT-4-based ICL. Six modules are included, all of which are intended to build context-aware knowledge graphs (KGs) from PubMed research articles. The process begins with a list of article titles that are supplied into reguloGPT.
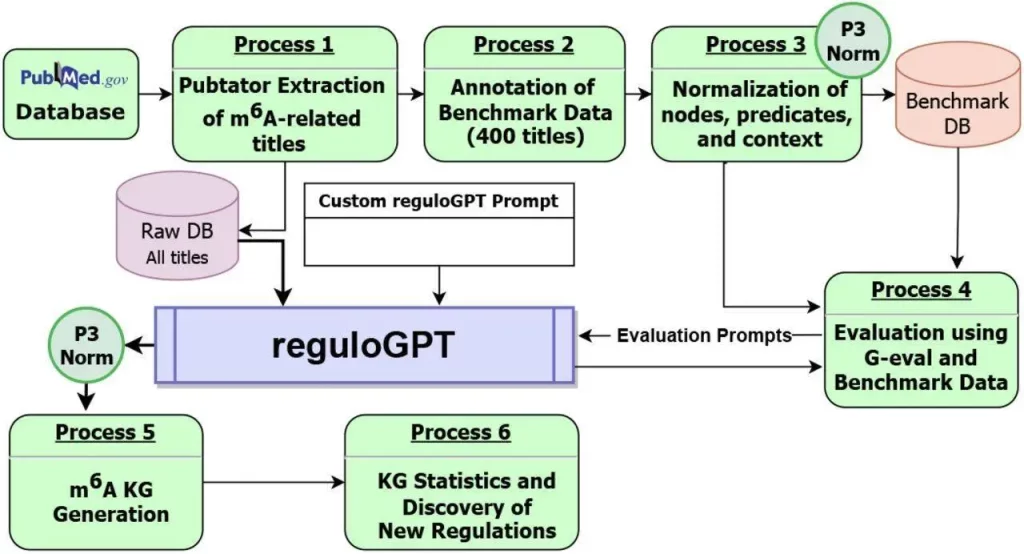
Image Source: https://doi.org/10.1101/2024.01.27.577521
Use of reguloGPT in Construction of m6A-KG
ReguloGPT was used by researchers to analyze 968 unannotated titles, and the output was context-aware relational graphs that show m6A-related functions in various scenarios. Researchers combine these relational graphs with annotated graphs from the benchmark dataset, normalizing the nodes, edges, and contexts to create a comprehensive m6A knowledge graph (m6A-KG), which represents the biochemical regulatory pathways associated with m6A. The built m6A-KG comprises 2,397 nodes, 4,694 edges, and 478 unique contexts. An average of 1.06 contexts are covered by each edge.
Conclusion
ReguloGPT is a novel approach introduced by researchers to make knowledge gaps in the context of MRPs. It is used by researchers to produce an extensive and thorough m6A-KG. The m6A-KG provided unique insights into the ways in which m6A functions in different types of cancer, leading to a more comprehensive understanding of m6A’s role in cancer and providing opportunities for focused cancer research and the development of targeted therapies. The effectiveness of the application is also determined by the researchers in this paper by producing notable results using a human-annotated benchmark database with 400 titles. These findings highlight the revolutionary potential of reguloGPT in terms of biological knowledge extraction from literature. In order to extract relationships from the KG in a methodical manner and conduct more productive research, future studies will enhance normalization schemes and clarify new regulatory roles. Subsequent research endeavors will delve into a more methodical evaluation of G-Eval and the extraction of relationships, in addition to enhanced normalization techniques for edges and contexts. Also, researchers will continue to establish a methodical and efficient strategy to clarify novel regulatory roles from the KG.
Story source: Reference Paper | The source code of reguloGPT, m6A title, and benchmark datasets, and m6A-KG are available on GitHub
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.