NovoMol is a novel de novo technique that increases the efficiency of clinical trial times by mass-generating therapeutic molecules with excellent oral bioavailability through the use of recurrent neural networks. The quantitative estimate of drug-likeness (QED) was used to rate the molecules after they were optimized for desired characteristics. Retraining the neural network using generated molecules that satisfied QED’s oral bioavailability criteria resulted in 76% of created molecules passing this stringent threshold and 96% passing the conventional Lipinski’s Rule of Five after five training cycles.
About Drug Discovery
The initial phase of drug development, known as drug discovery and development, involves finding potential drugs and referring them to clinical trials for additional research. This process reduces the available molecules to preclinical researchers, but despite technological advances, pharmaceutical research output has declined. The cost of FDA-approved medicines has increased 80-fold since 1950, leading to higher pharmaceutical costs, with prescription drug prices increasing by 6% and cancer medicine prices over 10%.
Virtual screening (VS) and high throughput screening (HTS) are efficient techniques for drug screening, although they can be laborious and have significant failure rates. These methods, which can take up to three months, frequently result in false positives, which reduces the pool of potential therapeutic candidates. Merely 3.4% of applicants for oncology and 13.8% of candidates for medications advance past the initial phase of clinical trials.
A promising method for creating therapeutic molecules from scratch using machine learning is called “de novo drug design.” Unlike current medication discovery approaches, this strategy can explore the chemical universe of feasible synthetic molecules and is not constrained by currently available chemicals. Nevertheless, it is frequently difficult for conventional computational methods like autoencoders and evolutionary algorithms to generate workable synthetic molecules with the required characteristics.
Introduction to NovoMol
NovoMol is an evolutionary de novo drug design method that uses a Recurrent neural network (RNN) to quickly and cheaply optimize for particular pharmacological properties. After synthesizing therapeutic candidates for a particular drug target using NovoMol, researchers evaluate the effectiveness of this strategy. Inspired by evolutionary methods, an RNN generates unique compounds in the form of SMILES strings. The top-generating molecules are then utilized to retrain the network via transfer learning. Researchers score produced medicines according to their molecular attributes using a new drug-likeness metric called Quantitative Estimate of Drug Likeness (QED). For FDA-approved oral bioavailable medications, the mean QED is 0.586. Specifically for orally accessible medications, NovoMol offers an objective-based approach to de novo drug discovery by optimizing molecules to remain above this barrier.
Understanding Recurrent Neural Network
Recurrent neural networks (RNNs) are used in natural language processing, and the study’s findings showed that over time, the neural network’s distinctive structure—many copies of the same neural network—can cause serious errors. This is a result of the data becoming diluted as it passes through hidden stages and is known as the “forgetting” problem, which might cause the model to overlook crucial information. For example, vanilla RNNs may fail to close branches when a set of brackets is used to indicate branching in SMILES molecules, resulting in invalid molecules. A long short-term memory (LSTM) recurrent neural network—which is capable of efficiently capturing and interpreting sequential and structured data—was used to solve this problem.
Using clusters of three neural networks, or cells, as opposed to simply one, LSTM networks are a kind of RNN created to address the “forgetting” problem. These cells play distinct roles in deciding which information is forwarded to the following cell and which must be kept in a state-designated for long-term storage. They are referred to as the forget gate, input gate, and output gate. Because SMILES molecules have this property, LSTMs can retain important details in long-term memory, which improves the accuracy of their model.
The study makes use of the Python, TensorFlow, and Keras frameworks to create an encoder-decoder neural network with an LSTM, input, and dense layers. The encoder reduces the dimensionality of the data while retaining pertinent information by converting each character of the input, which are 100-character long SMILES molecules, into a latent representation. The original SMILES molecule is read in compressed format, and the decoder attempts to reconstruct it. During training, the reconstruction error—that is, the discrepancy between the original and decoded molecules—becomes important. The goal of both the encoder and the decoder is to determine the best compression and molecular reconstruction technique.
Training NovoMol
The model was developed using the ChEMBL Python API to filter out drug molecules with a QED value greater than or equal to 0.586 that are orally administered. Oral bioavailability is crucial in drug development, as oral drugs are safer and cheaper. The model was retrieved in SMILES format, which uses ordered characters to represent the overall chemical structure of molecules. Unique characters represent atoms, bonds, and rings, allowing SMILES strings to store molecular information. The model then added start (!) and stop (E) characters to each SMILES molecule, increasing the unique character count to 45 characters.
To differentiate between molecules, stop characters (E) were padded to a length of 100 characters. SMILES molecules were one-hot encoded, representing each character as a 45-dimensional vector of zeros with ones in the character index, allowing the model to easily learn patterns in the retrieved molecules.
With 482,000 SMILES molecules used for training, the entire model produced very little reconstruction error. Three LSTM layers, an input layer, and a dense layer that condensed hidden and cell states made up the model. An input layer, dense layers, LSTM layers, and a dense layer make up the decoder. The Adam optimizer and backpropagation were used to train the network. On a Google Colab Tesla P100 GPU, the model was trained for 225 epochs, yielding 99.96% training accuracy and 99.86% test accuracy. To produce novel molecules, a model that acts as an intermediary was incorporated between the encoder and decoder models. The model produced molecules with increased diversity, maintaining its excellent accuracy.
Future Prospects
NovoMol is expected to be improved using additional AI techniques like reinforcement learning, as well as by introducing novel scoring systems to motivate the model to produce progressively more varied and advantageous molecules in the future. The model will also be extended to provide reaction-based de novo design for molecule creation and property prediction using machine learning techniques and structural warnings for virtual screening.
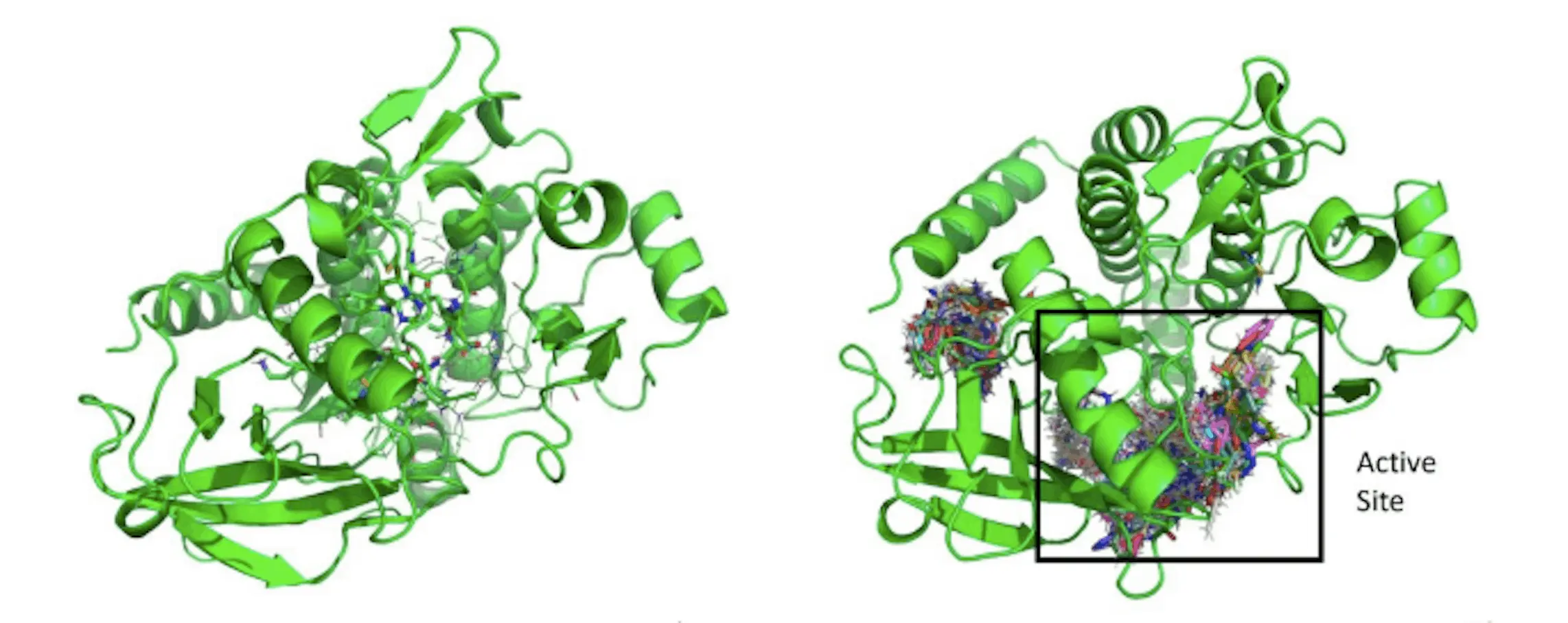
Validating NovoMol’s efficacy will also involve testing for a number of additional pharmacological targets. EGFR (lung cancer), HER2 (breast cancer), HIV-1 RT (HIV/AIDS), and AChe (Alzheimer’s illness) are a few possible targets. A growing number of researchers working on drug development pipelines are choosing to employ in-silico de novo drug design techniques. Current drug development schedules and costs could be lowered with the use of a flexible and precise method like NovoMol. Researchers are looking forward to NovoMol as it may speed up the development of new therapeutics and bring these essential drugs to the market sooner by producing higher-quality drug candidates.
Conclusion
NovoMol is a successful approach that uses a recurrent neural network with long short-term memory (LSTM) to generate an orally bioavailable de novo drug design. The model optimizes molecular properties for high QED by generating distinct and diverse molecules with similar properties. After five iterations, molecules with higher average QED were obtained by iterative improvement of the model. NovoMol has undergone a retraining process to create drugs targeting the oncoprotein PDGFRα, which will hopefully result in fewer side effects and more alternatives with improved pharmacological characteristics. Additional laboratory experiments are required to confirm these features and additional impacts.
Article Source: Reference Paper
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.