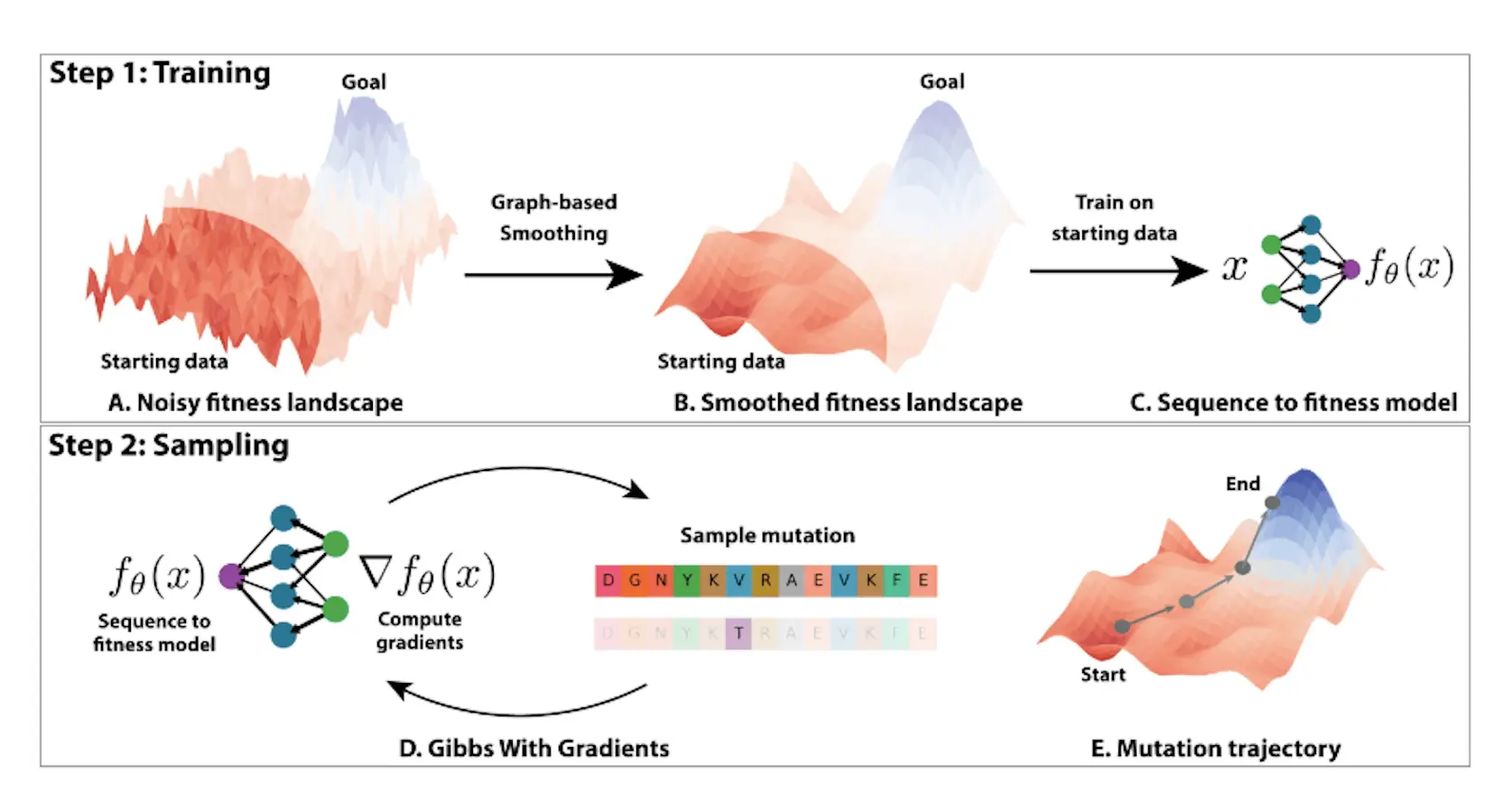
Modeling the combinatorially huge number of sequences is not practical since previous approaches frequently restrict optimization to a narrow mutational radius, restricting the design space for novel proteins with improved fitness for a desired attribute. Researchers from the Massachusetts Institute of Technology suggest flattening the fitness landscape in place of heuristics to make protein optimization easier. Protein fitness is first expressed as a graph signal, and the fitness landscape is then smoothed using Tikunov regularisation. In the Green Fluorescent Proteins (GFP) and Adeno-Associated Virus (AAV) benchmarks researchers find that optimizing in this flattened landscape improves performance across a variety of techniques. Second, researchers use MCMC in the smoothed landscape and discrete energy-based models to get state-of-the-art results. With in-silico evaluation, the researchers’ method—Gibbs sampling with Graph-based Smoothing, or GGS—shows a remarkable capacity to get a 2.5-fold gain in fitness over its training set. GGS shows promise for protein optimization in the low-data setting.
Introduction
Protein engineering focuses on improving fitness by enhancing performance on a desired property or function. Starting with protein optimization, the number of possible proteins increases exponentially with sequence length, making brute-force search infeasible. Directed evolution has been successful in improving protein fitness but requires substantial labor and time. Examples include catalytic activity for enzymes and fluorescence for biomarkers.
The objective is to maximize a learned model of the fitness landscape to produce high-fitness proteins. Nevertheless, non-smooth fitness landscapes, experimental noise, and zero fitness present problems for proteins. Due to the scarcity and high cost of protein fitness datasets, machine learning techniques are prone to producing false positive results and being trapped in local optima. A smoothing regularizer is presented to predict similar sequences to have similar predicted fitness in order to address these problems. Smoothing, particularly in discrete optimization, can be a useful tool since it helps gradient-based approaches avoid local optima and attain higher peaks. Smoothing within a graph framework has not been directly implemented in many research, although it is a useful technique for navigating the noisy fitness environment and enhancing protein performance.
Key Features of the Study
- Researchers created GGS, a novel sequence-based protein optimization approach that trains a smoothed fitness model via graph-based smoothing. In order to progressively sample mutations towards higher-fitness sequences with GWG, the model is employed as a discrete energy function.
- Researchers create a series of activities that assess how well a strategy may generalize to greater fitness levels. In order to simulate challenging optimization scenarios where limited and noisy data are initially accessible, researchers employ publically available GFP and AAV datasets.
- Researchers’ benchmark demonstrates that earlier techniques cannot be extended to higher fitness levels. Graph-based smoothing, on the other hand, can significantly boost their performance; in one baseline, the fitness increases from 18% to 39% in GFP and from 4% to 44% in AAV following smoothing.
- In comparison to the next best method, this method, GGS, achieves state-of-the-art outcomes with five times higher fitness in GFP and two times higher in AAV by directly exploiting smoothness.
Introducing Novel Method to Improve Fitness Landscapes
The paper suggests a novel way to use the graph Laplacian as an optimization tool to apply smoothing to protein sequence and fitness data. Fitness values are used as node attributes in the formulation of the sequences as graphs, and Tikunov regularisation is used to smooth the topological signal. For discrete optimization, a neural network is fitted to the data. High fitness sequences are sampled over the energy function using the Gibbs With Gradients (GWG) technique, which creates a discrete distribution based on the gradients of the model. An iterative sampling of subsequent mutations follows, directing the population towards increased fitness.
Previous Related Studies
Protein optimization and design – The problem of optimizing a noisy fitness landscape is faced by sequence-based techniques such as model-based directed evolution, generative models, and reinforcement learning. The research is centered on sequence-based techniques that make use of Gibbs With Gradients (GWG), which achieves cutting-edge results in discrete optimization but demands a smooth energy function to perform well. GWG was utilized by Emami et al. (2023) to optimize proteins, but the outcomes were not up to par. The study improves on previous approaches by addressing the requirement for better regularisation and smoothing in sequence-based algorithms.
Protein fitness regularization – An early attempt to use a statistical model of epistasis to model protein fitness smoothness was the NK model. A generalized NK model was presented by Brookes et al. (2022) as a framework for approximating protein fitness sparsity. A comparable technique called dWJS regularises the discrete energy function during Langevin MCMC by using Gaussian noise. Iterative optimization is meant to level the fitness landscape so similar sequences fit equally. Instead of analyzing various smoothing techniques, the emphasis is on highlighting how crucial they are to training.
Limitations
- The outcomes provide compelling support for the use of smoothing, considering its advancement in several areas. Despite this, the assessments adhere to previous research by employing a trained model for assessment. When compared to testing sequences using wet-lab validation, this can be inconsistent. Wet-lab validation, regrettably, can be expensive and time-consuming. Using GGS in an experimental or active learning pipeline with wet-lab validation integrated into the loop would be the ultimate test.
- Finding the ideal hyperparameters for proteins requires the use of hyperparameters such as graph size and smoothing parameter γ. Although researchers’ selections of hyperparameters are not exclusive to AAV or GFP, they can be applied to estimate the fitness landscape. The problem of protein optimization can be advanced by utilizing the linkages between spectral graph theory and protein optimization, which can lead to a more efficient method of protein optimization.
Conclusion
Graph-based Smoothing (GGS) is a new graph signal processing technique that combines a smoothed fitness landscape and Gibbs sampling for protein optimization. This method enhances the fitness landscape as a whole and baseline performance. The combination of Gibbs and gradient with a smoothed model yielded the best results for GGS, indicating the beneficial effects of gradient-based sampling when combined with a smooth, discrete energy-based model. The outcomes underline the advantages of optimizing as opposed to a smooth landscape, which might not be a real representation of the fitness landscape. Instead of learning too much biological noise, the objective is to identify the signal in the data to determine which protein is optimal. In order to convert protein fitness data into a format that is compatible with current optimization algorithms, regularisation is essential.
Article source: Reference Paper | Reference Article | GGS code is available on GitHub
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.