Protein engineering is in the middle of a quiet revolution, and a new study from researchers at the School of Computational Science and Engineering, Georgia Institute of Technology, pushes that revolution a big step forward, introducing SPURS (stability prediction using a rewired strategy). Their work, published in Nature Communications, tackles a longstanding challenge in computational biology: how to predict the impact of amino acid substitutions on protein thermostability in a way that is both accurate and scalable enough for real-world design and disease interpretation. By cleverly rewiring two powerful protein generative models, they turn abstract sequence and structure priors into a practical engine that can profile mutation effects across entire proteomes.
Why Protein Stability Still Matters
Changes in Gibbs free energy (ΔG) can help define thermodynamic stability, which is one of the many parameters that can help describe if a protein folds, functions, or fails. Deep mutational scanning and directed evolution can describe parts of the protein landscape, but they are costly, biased toward destabilizing variants, and limited to only a few proteins. Classical machine learning models were helpful, but were trained on modest and heterogeneous datasets, which restricted the models’ generalizability to unseen proteins and rare stabilizing mutations. Conversely, large protein language models and inverse folding models were not designed for supervised stability predictions, but had strong “zero-shot” correlations with stability and fitness.
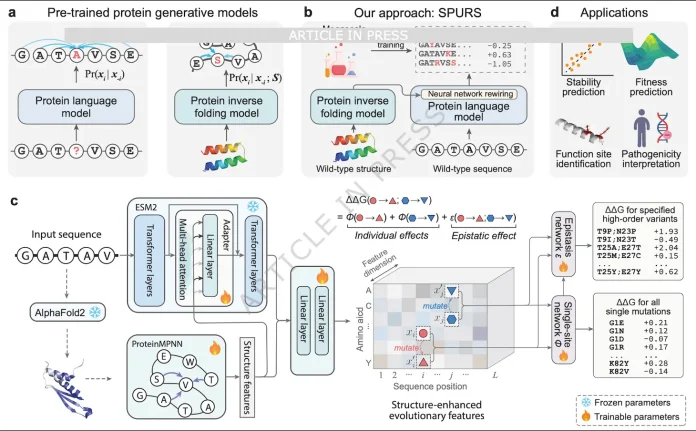
SPURS: Rewiring Sequence and Structure
SPURS operates on the principle that it is more useful for sequence models and structure models to be collaborative rather than adversarial. The architecture utilizes a wild-type protein sequence and its associated 3D model. The sequence is encoded with evolutionary information using ESM2, while the 3D geometry is encoded using ProteinMPNN. Rather than simply stacking the outputs, researchers use a technique with a sequence of ESM2 that is enhanced with a ProteinMPNN Embedding stack to form a better evolutionary representation. ESM2’s 650M parameters are frozen, and only the Adapter plus ProteinMPNN (about 9.9M parameters) are fine-tuned, cutting trainable parameters by ~98.5% compared with full pLM fine-tuning and greatly reducing overfitting risk.
Benchmarks: From Megascale to Human Proteome
Researchers trained SPURS on the Megascale dataset, which includes over 770,000 stability measurements across 479 domains, and features dense coverage of single and selected double mutants. SPURS has a median Spearman correlation of ~0.83 on the Megascale test set, with the most out of any proteins when compared to ThermoMPNN, with the exception of stabilizing mutations, which most models fail to identify. SPURS achieves performance that is at least on par with the current state-of-the-art methods, including FoldX, Rosetta, ThermoNet, Stability Oracle, and ThermoMPNN, across 12 distinct datasets, as measured by ΔG or melting temperature. SPURS still outperformed ThermoMPNN on the large Human Domainome dataset (over half a million human variants), improving the correlation with stability-related abundance phenotypes, demonstrating that the improvement still stands at the proteome level.
Beyond Stability: Function, Fitness, and Disease
The most fascinating fact about SPURS is how smoothly it integrates with other analyses. Even though SPURS was not trained on functional annotations, the authors, by combining SPURS-predicted ΔG with ESM1v fitness scores via a sigmoid fit and residual analysis, craft a per-residue function score that pinpoints binding regions, catalytic sites, zinc-coordination sites, and other hotspots. Adding SPURS ΔGΔG as a simple extra feature to established low-N fitness models, such as Augmented DeepSequence, yields consistent improvements across 141 deep mutational scanning datasets, especially for expression and organismal fitness. At the edge of the clinic, SPURS shows that, on average, the destabilizing pathogenic variants are more destabilizing than benign ones, and the less solvent-exposed they are, the more buried, folded regions. And ΔG and solvent exposure combination separates pathogenic from benign variants, achieving an AUROC of ~0.84.
Looking Ahead: SPURS as a Stability Prior
Taken together, SPURS feels less like yet another predictor and more like a stability prior that can be wired into many pipelines in protein informatics. SPURS’s proof of concept shows that, through the careful reconfiguration of multi-modal generative models, we can achieve a superlative level of performance, scalability, and efficiency, all without the need to train a new model from the ground up. For protein engineers, it provides a high-throughput way to enumerate and rank combinatorial mutational degenerate libraries; for geneticists, it offers a perspective to dissociate stability-driven pathogenicity from other mechanisms that are associated with altered interactions and/or gain-of-function. As the community shifts toward generative design, models such as SPURS will productively serve as reward functions, filters, and guiding fields to ensure that the sequences designed by A.I. are sufficiently novel but also sufficiently stabilized to be of use.
Article Source: Reference Paper | Source Code: GitHub
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.