Predicting the immunogenicity of peptide antigens attached to major histocompatibility complex (MHC) molecules is critical for developing new immunotherapies and better understanding human immune responses. While existing techniques rely on simple sequence representations, they neglect the complex chemical mechanisms that drive peptide recognition. Researchers from Cleveland Clinic and IBM collaborated to create supervised and unsupervised AI to expose the molecular properties of peptide antigens, which are tiny fragments of protein molecules that immune cells employ to identify threats. Project participants were from a variety of groups supervised by Cleveland Clinic’s Timothy Chan, M.D., Ph., as well as IBM’s Jeff Weber, Ph.D., Senior Research Scientist, and Wendy Cornell, Ph.D., Manager and Strategy Lead for Healthcare and Life Sciences Accelerated Discovery. Published in Briefings in Bioinformatics, their findings give light on the intricate interplay of peptide structure, kinetics, and MHC interactions that influence T-cell recognition. This greater understanding has enormous implications for refining T-cell based immunotherapies and developing more effective therapeutic T-cell receptors.
Introduction
The immune system’s ability to recognize and kill foreign invaders is dependent on its ability to identify peptide antigens presented by MHC molecules on cell surfaces. This identification induces T-cell activation, a key stage in adaptive immune responses, and recognition is based on the presentation of peptide antigens linked to MHC molecules on cell surfaces. Accurately forecasting the immunogenicity of MHC-peptide complexes is thus critical for creating successful immunotherapies for cancer and other illnesses. Traditional methods for estimating immunogenicity are mainly reliant on peptide sequencing. While these approaches provide some insight into immunogenic aspects, they only capture some of the complexity of peptide-MHC interactions, which include subtle structural and dynamic features.
Scientists have spent decades exploring how to better identify antigens and use them to assault cancer cells or virus-infected cells. This task has been difficult because antigen peptides interact with immune cells based on unique properties on their surface, a process that is still not fully understood. The sheer number of variables that influence how immune systems recognize these targets has hampered research efforts. Identifying these variables is difficult and time-consuming with regular computation; therefore, existing models are limited and occasionally wrong.
Challenges in Predicting Immunogenicity
- Current AI models rely on text-based amino acid sequences.
- These models are ineffective with short datasets and cancer-derived antigens.
- They do not capture structural and dynamical characteristics important for TCR binding.
Researchers from Cleveland Clinic and IBM have published a strategy for identifying new targets for immunotherapy through artificial intelligence (AI), which showed that unsupervised AI can detect small differences in the immunogenicity of a cancer neoantigen against its wild-type equivalent. Furthermore, a supervised AI model beats sequence-based approaches in categorizing immunogenic peptides, even when trivial sequence correlations are taken into account. Notably, both unsupervised and supervised techniques identify critical immunogenicity factors outside the MHC binding groove, such as time-dependent molecular fluctuations and anchor position dynamics.
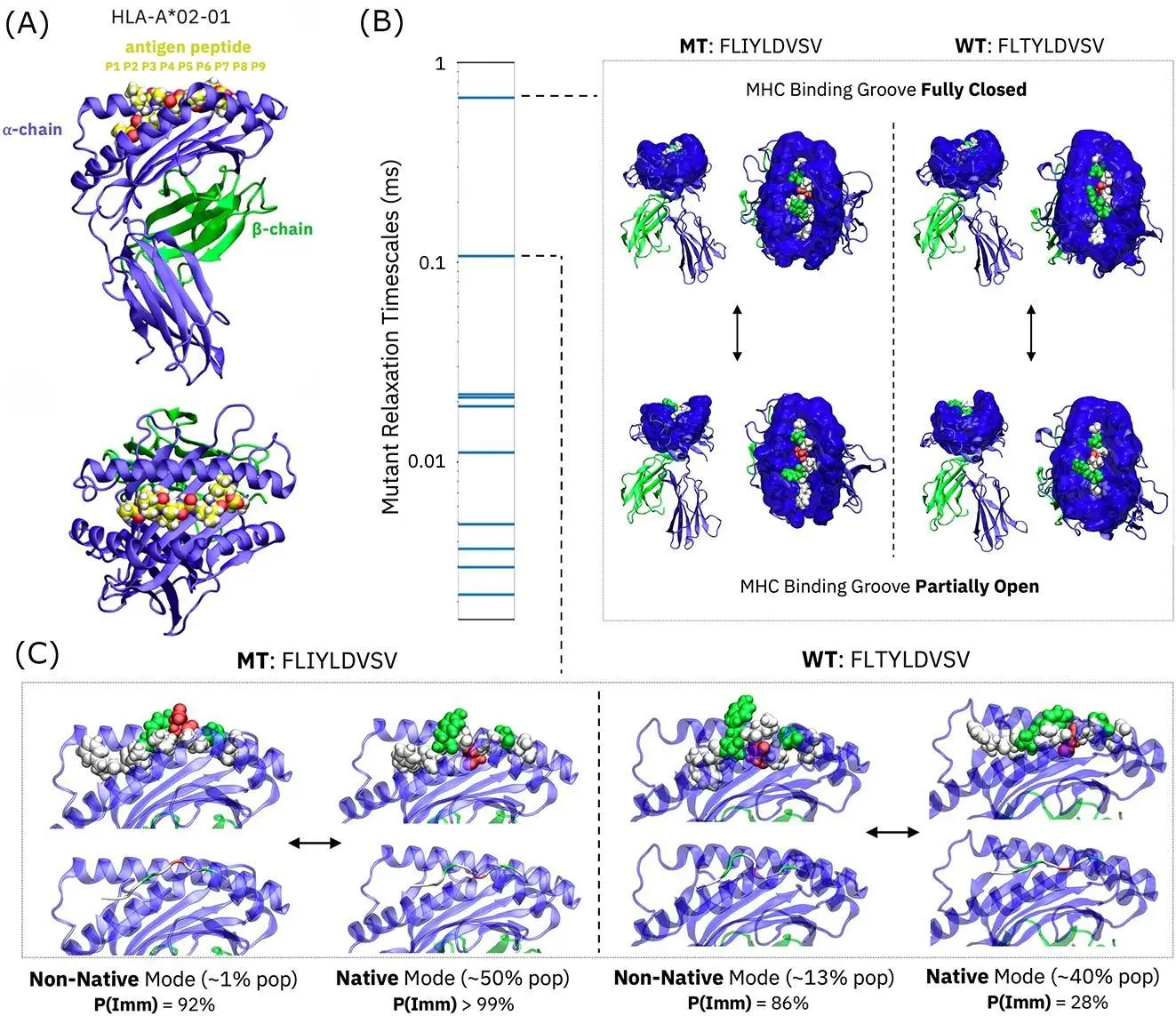
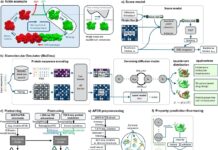
They used artificial intelligence to explore deeper into the factors that influence the immunogenicity of the MHC(HLA-A2)-peptide complex, using unsupervised and supervised AI techniques on massive datasets derived from molecular dynamics simulations of peptide-MHC interactions.
They used the following AI approaches:
- Unsupervised Markov Models
- Estimate slow molecular kinetics and create new MD trajectories.
- Capture processes that extend beyond the timeframes of individual MD simulations.
- Supervised molecular graph convolutions
- Integrate local molecular interactions into multiscale structural representations.
- Use physical potential energy functions to enhance categorization.
Methods
Molecular dynamics simulations:
- All-atom MD simulations of HLA-A2 bound to immunogenic and non-immunogenic peptides (training and testing datasets).
- Researchers used the AMBER16SB force field and the TIP3P water model with explicit solvent.
- Equilibrated simulations lasting 100 ns each produced conformational ensembles for each peptide-MHC complex.
- Extracted different structural and dynamic aspects from the trajectories, such as:
1. Cα locations and root mean squared fluctuations (RMSF)
2. Interatomic Distances and Angles
3. Hydrogen bond networks
4. Potential Energies
Unsupervised Learning using Markov Models:
- Hidden Markov models (HMMs) were built from MD trajectories to represent long-range conformational dynamics.
- Dimensionality reduction techniques (t-SNE and UMAP) were used to depict the HMM-identified microstates.
- Differences in microstate occupancy between immunogenic and non-immunogenic peptides were compared.
Supervised Learning using graph convolutional networks (GCNs):
- Peptide-MHC complexes were represented as molecular graphs, with nodes corresponding to atoms and edges indicating interactions.
- GCNs were used to learn multi-scale graph representations, which included structural and dynamic information from MD simulations.
- GCNs have been trained to distinguish peptides as immunogenic or non-immunogenic using previously learned representations.
- Model performance was measured using criteria like accuracy, precision, recall, and F1-score.
Data correction for trivial sequence correlations:
- To account for potential biases caused by sequence homology, the researchers removed substantially similar peptide sequences from the training dataset.
- This stage guaranteed that the GCN model learned generalizable characteristics based on structural and dynamic qualities rather than simple sequence similarities.
Keys Findings
- Recognition of peptide antigens on MHC molecules plays a crucial role in immunotherapy and human health.
- Current approaches for estimating antigen peptide immunogenicity are insufficient, as they only evaluate simple sequence representations.
- AI algorithms can predict the immunogenicity of the HLA-A2 peptide complex.
- Unsupervised AI can find small changes in immunogenicity between cancer neoantigens and wild-type peptides.
- Supervised AI approaches outperform sequence models for predicting MHC (HLA-A2)-peptide complex immunogenicity.
- Both unsupervised and supervised AI approaches can discover determinants of immunogenicity based on time-dependent molecular fluctuations and anchor position dynamics outside the MHC binding groove.
- These findings have ramifications for the generation of T-cell responses and the therapeutic T-cell receptor design.
Conclusion
Understanding how immune cells sense external invaders is critical to creating effective immunotherapies. This recognition is based on the presentation of peptide antigens linked to MHC molecules on cell surfaces. Researchers investigated artificial intelligence (AI) as a technique for predicting and understanding the immunogenicity of these MHC-peptide complexes.
The combined AI and simulation approach can highlight small but important determinants of peptide immunogenicity within the MD trajectory data and provide much greater prediction power than a baseline sequence architecture on peptide datasets. Markov models are initially used to investigate the role of conformational structure and dynamics in immunogenicity. Classification models are then created using large-scale MD datasets containing thousands of peptidic antigens. Low-data conditions that adjust for trivial sequence correlations result in improved classification performance. Making predictions using tiny training datasets is sometimes the sole option for investigating cancer neoantigens and lesser-studied HLA alleles. The researchers’ MD-AI results indicate additional pathways of peptide immunogenicity related to peptide anchor dynamics and peptide fluctuations in general. These findings illustrate how MD can assist in anticipating and explaining immunogenicity, and the methodologies established here provide the groundwork for large-scale HLA allele investigations to elucidate immune response pathways and inform T-cell treatments.
These findings provide new potential in immunotherapeutic development. Understanding the dynamic drivers of immunogenicity allows us to:
- Choose highly immunogenic peptides for vaccines and T-cell treatments with higher precision.
- Create more effective T-cell receptors that address the dynamic features of peptide-MHC interactions, resulting in stronger immune responses.
- Create innovative immunotherapies that go beyond static concerns, broadening the scope of therapeutic possibilities.
Article Source: Reference Paper | Reference Article
Follow Us
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.