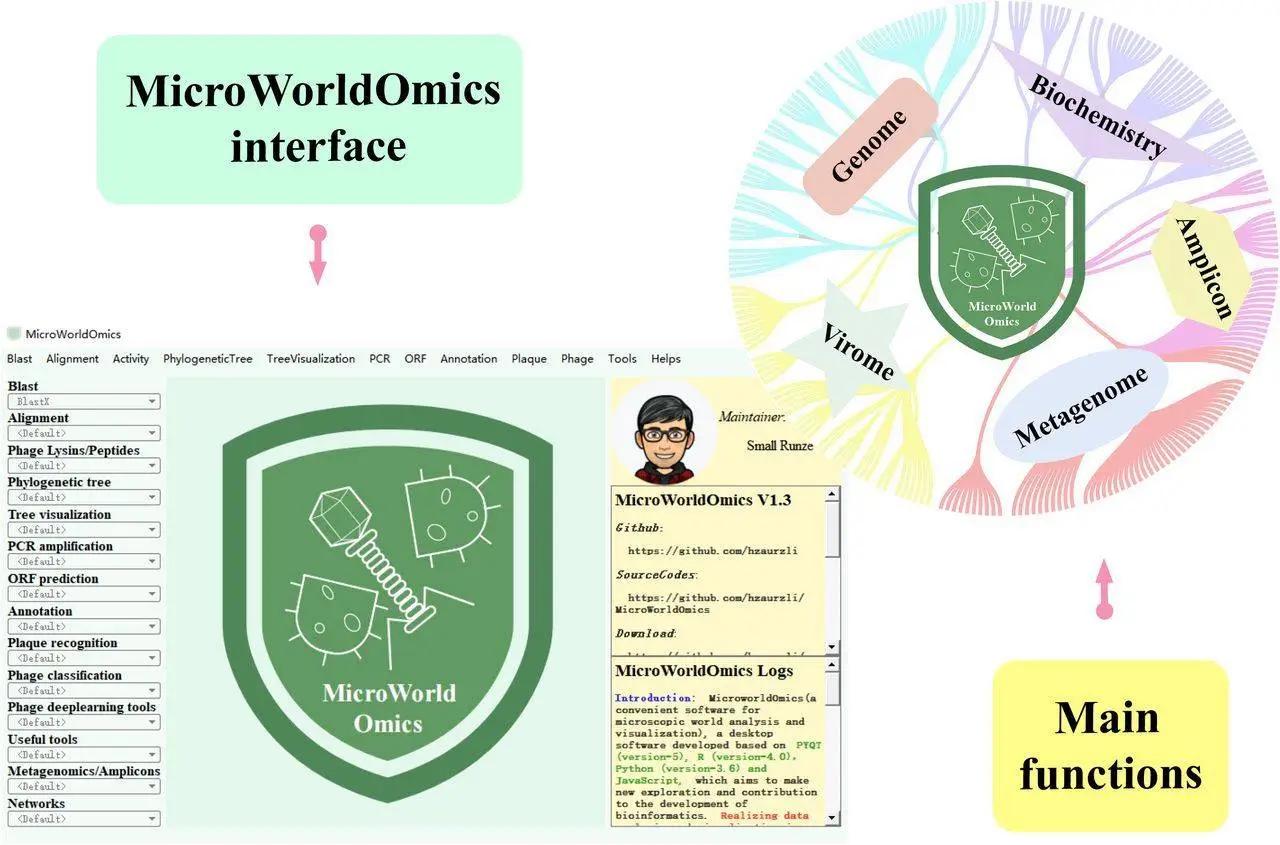
Dr. Runze Li and his colleagues from different institutes in China have recently developed high-tech tools that help researchers navigate through the complexities of the microbial world. MicroWorldOmics is a powerful desktop app that aims at simplifying and optimizing the analysis of microbiomes and viromes.
The body is an intricate and delicate ecosystem inhabited by countless microbes, including bacteria, archaea, or viruses (phages). These are called the microbiome and virome, which play a significant role in our health and well-being. However, it may be so hard to comprehend the secrets of this “micro world.” In this blog, we will look into MicroWorldOmics, the functionalities offered by its rich graphical interface, and how it helps you as a researcher to make breakthroughs in this exciting scientific field.
MicroWorldOmics: A Comprehensive Toolkit for Microbiome and Virome Exploration
Imagine a software suite that helps you analyze microbiome and virome data, with or without coding skills. This is the magic of MicroWorldOmics! Runze Li and his team have developed this desktop application, which has several features to fast-track your research.
User-Friendly Interface: Forget about complicated command lines. For ease of navigation, MicroWorldOmics comes with a visually appealing graphical user interface. Input, output, parameters, and status are all clearly partitioned.
Plugin Powerhouse: More than 90 plugins divided into 14 modules are at your disposal! MicroWorldOmics comes with over ninety plugins divided into fourteen categories. It does not matter if you want to make phylogenetic trees understand plaque signatures or learn more about that microbial “dark matter” later on; there is a plugin for that.
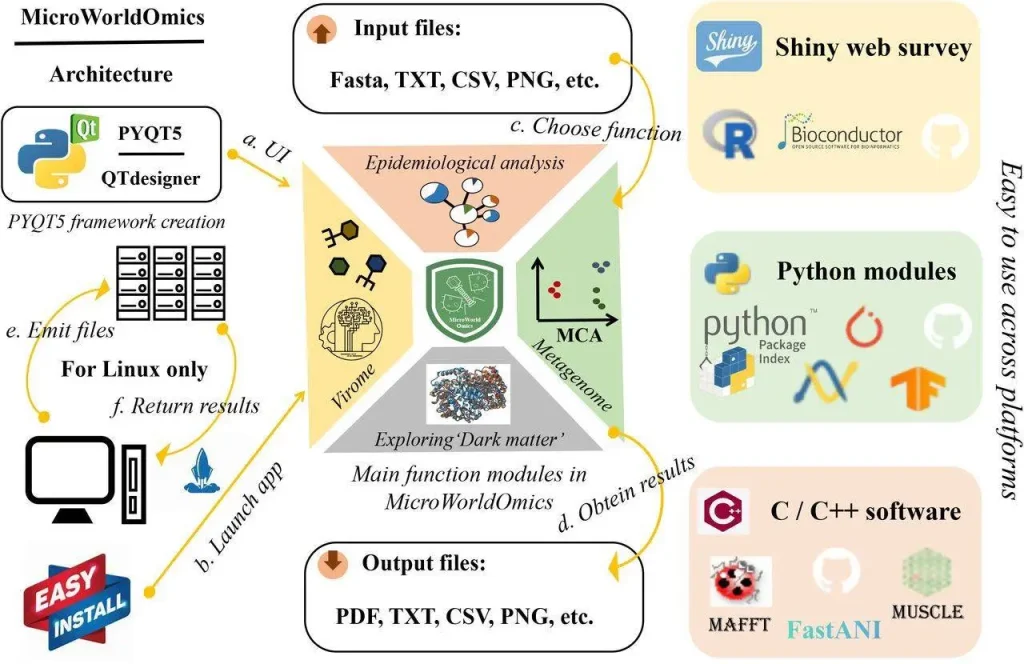
Platform Agnostic: Operating systems should be the least of your concerns. MicroWorldOmics can run smoothly on Windows, Linux, and Mac machines for researchers using different platforms.
Data Visualization and Steroid Effects: Gone are the days of static data tables. Shiny apps, which are easily incorporated with MicroWorldOmics, allow you to visualize and analyze your data interactively. Generate amazing graphs, heat maps, and network diagrams to understand better microbes in our environment.
A variety of Formats for Data Flexibility: You should not be worried if you have different formats of data. TXT, CSV, PDF, JPEG, or PNG files can all be supported by MicroWorldOmics software. Simply import your data and start doing real analysis.
Sample Data Playground: Do you feel uncomfortable about a particular plugin? Worry no more! These sets of sample data provided by this software help the user learn how to use it and refer back for any clarifications needed.
Microbiome and Virome Unveiled
MicroWorldOmics provides a versatile toolkit for microbiome and virome analysis. Let’s look further into some cool applications that the software enables:
Epidemiological Investigations: Studying the spread of pathogenic bacteria? In this regard, MicroWorldOmics has a range of plugins. These include analyzing multi-locus sequence types (MLST), detecting serotypes, and identifying drug-resistance genes in one platform. Interactive PCA plots show large-scale isolate genomic distances to help elucidate how strains of bacteria change and are transmitted.
Virome Analysis with Artificial Intelligence (AI): Phages, the most prevalent viruses on earth, play an important role in the microbial ecosystem. By using AI, MicroWorldOmics helps unravel the secrets behind viromes. This involves finding phage sequences and taxonomical classification, as well as distinguishing between lysogenic or lytic life cycles of phages and predicting their hosts by employing state-of-the-art machine learning algorithms.
Genome-Based and Metagenome-Based Analysis: MicroWorldOmics caters to isolate- and community-level analysis. Whole genome sequencing data can be analyzed to reveal finer details about individual isolates, such as worldwide distribution patterns, evolutionary relationships among them, and genes involved in island hopping, for example. On the other hand, metagenomic or amplicon sequencing data can provide broader insights into the composition and functioning of entire microbial communities.
Visualizing Microbial Networks: MicroWorldOmics goes beyond static data analysis towards dynamic microbial network visualization. It enables you to construct and visualize complex microbial networks. Different network types, such as correlation-based, graphical models, and differential networks, allow an understanding of the intricate web of interactions within the microbial ecosystem. Network features can be analyzed to identify key microbes associated with specific conditions.
Exploring the “Dark Matter” of the Micro World
A significant portion of the microbial world remains uncharacterized, often referred to as the “dark matter.” This vast unknown is acknowledged by MicroWorldOmics, which has functionalities meant to explore this uncharted territory. This software integrates diverse bioinformatics tools and algorithms that can help researchers detect and analyze these mysterious microorganisms, a process that may ultimately lead to groundbreaking discoveries.MicroWorldOmics simplifies the exploration of the microbial universe.
Join the Conversation!
MicroWorldOmics is a powerful and ever-evolving platform. Share your thoughts and experiences with MicroWorldOmics. Did you find this blog informative? Comment to let us know!
Article Source: Reference Paper | The development details and download address of “MicroWorldOmics” are available on GitHub.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.