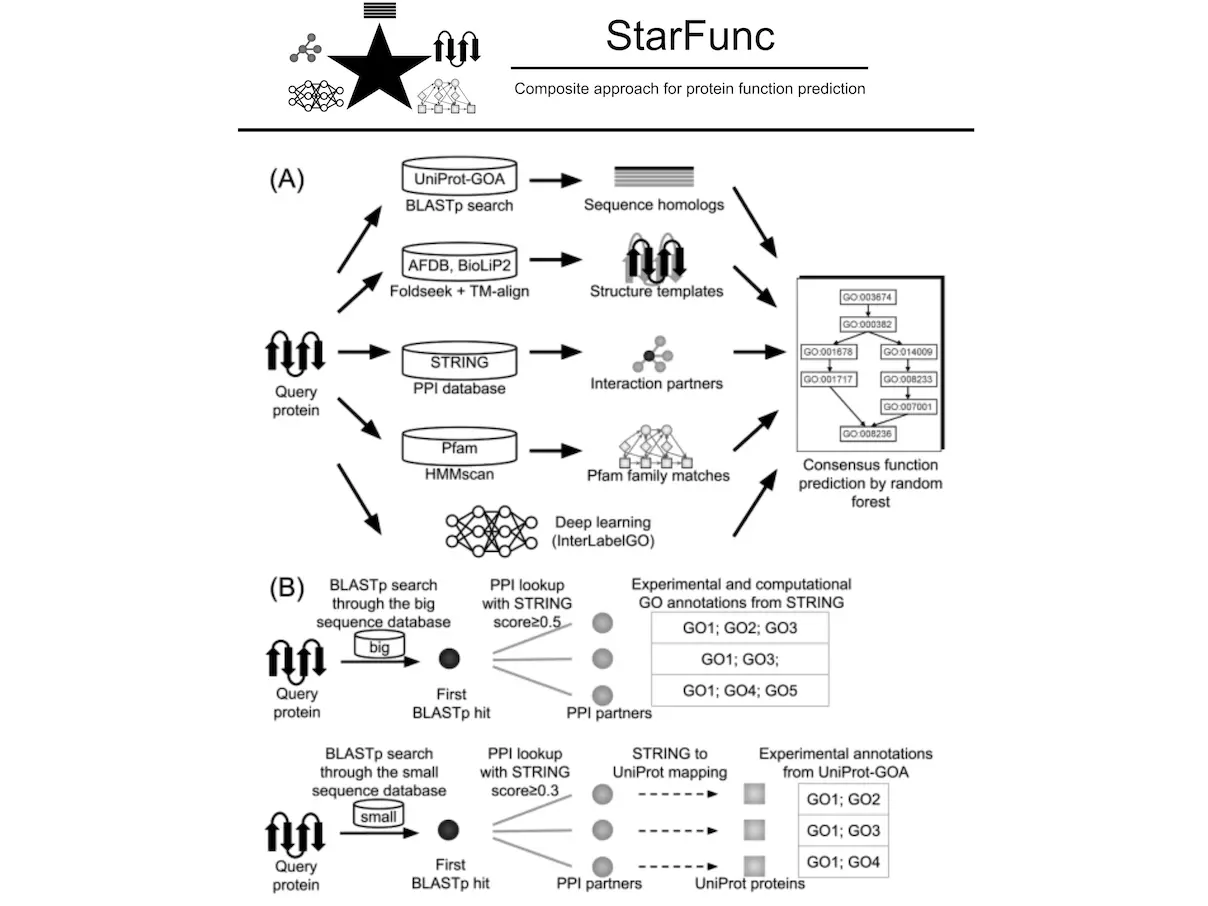
A recent study by the Chengxin Zhang lab from the University of Michigan introduced StarFunc, an accurate protein function prediction approach integrating cutting-edge deep learning with template data from sequence homology, protein interactions, structural similarities, and domain families. Rather than picking sides, however, StarFunc combines two worlds to create a unified framework. This creative idea is expected to transform the field of protein function prediction and usher it into an era of accuracy and efficiency.
Imagine tiny machines that make our cells work, proteins! Each protein has a specific function, ranging from building structures to complex chemical reactions that take place within them. Protein functions thus describe their roles, which are essential in unraveling health mysteries, diseases, or drugs. Nevertheless, experimental determination of protein functions can be like searching for a needle in a haystack, consuming time and resources.
Generally, these methods have been based on either templates or deep learning. Template-based approaches compare unknown function proteins (target proteins) to proteins that have known functions and structures. On the other hand, deep learning is capable of recognizing hidden patterns within large datasets containing protein sequences and functions but only sometimes uses available information about structures.
StarFunc: A Multifaceted Approach in Predicting Protein Function
This approach combines five different methods of predicting protein function:
- Sequence-based annotation: This entails comparing the amino acid sequence of the target protein to other characterized proteins. When sequences are similar, functions are likely to be similar too.
- Structure-based annotation: In this case, StarFunc maps the 3D structure of the target protein with known structures that have known functions. Proteins with similar structures are more likely to have similar functions.
- Protein-protein interaction (PPI) based annotation: Most proteins do not function alone. This technique identifies interacting proteins and deduces their role from those.
- Pfam-based annotation: The Pfam database groups proteins into families based on their sequence and structure. For example, if there are related members within a family, then they may share functions. This is what StarFunc makes use of in predicting.
- Deep learning–based annotation (InterLabelGO): This unit is powered by a deep learning model that has been trained on a large dataset consisting of protein sequences and their corresponding functions. Thus, it can directly predict a protein’s function from its sequence alone.
StarFunc’s Success
The secret of StarFunc lies in its ability to incorporate these different techniques. StarFunc uses sequence similarity, protein structure, interaction partners, protein families, and deep learning to achieve far more accurate results than single-method approaches.
This research has shown that StarFunc consistently outperforms all individual methods as well as other latest algorithms in predicting:
- Molecular Function: What protein does at the molecular level (e.g., breaking down sugars, building muscle)
- Biological Process: The broader cellular role of the protein (e.g., energy production, cell signaling)
- Cellular Component: Where in the cell the protein functions (e.g., cell membrane, nucleus).
Another added advantage of StarFunc is efficiency. The study also compared it with COFACTOR, which is an older structure-based prediction method. It delivers much faster predictions compared to COFACTOR and does so while maintaining a high level of accuracy. Imagine getting the same or perhaps better results within less time – this is how powerful StarFunc can be!
The Future of StarFunc’s Horizons
Though it is a very big step forward by StarFunc, the authors still admit there is room for improvement. One area that deserves exploration in this regard is text mining. In recent times, some research studies have shown that analyzing scientific literature together with protein sequence and structure data can further enhance such predictions. Imagine what would happen if StarFunc took into account the latest scientific findings in its predictions!
Moreover, the deep learning constituent of starfunc could be enhanced by incorporating more features emanating from sequence alignments and protein structures; such features could help deepen the model’s understanding of proteins, thereby leading to more accurate predictions.
These shortcomings will be addressed in future versions regarding StarFunc. They also hope that integrating text mining with additional features will improve its performance and robustness, making it one of the most powerful tools for predicting protein function.
Start a Conversation!
The combination of various prognostication methods by StarFunc marks the dawn of a brand new chapter in protein function prediction. It also allows for a deeper understanding of biological systems and protein-protein interactions and leads to the discovery of new drugs and therapies. In this regard, StarFunc has the potential to significantly speed up biological research and reveal scientific truths.
Hang on! The discussion does not end there. We would like to hear from you:
What is your opinion on how StarFunc combines disparate methods? Let us hear your ideas and start a conversation!
Article Source: Reference Paper | StarFunc is available on the Zhang Lab’s Webserver.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.