Researchers from the Institute of Bioengineering, Lausanne, Switzerland, introduce GEMLI, a powerful computational tool that enables robust identification of cell lineages solely from scRNA-seq data. By leveraging gene expression memory, GEMLI unveils heritable patterns, cell fate decisions, and multicellular structures, shedding new light on processes like cancer invasiveness.
Introduction
What if you have to take a snapshot of a busy city during rush hour? Back and forth, vehicles move with their destinations. Can you tell which vehicles set off at the same time? It’s what scientists studying cell lineages have to face: the family trees of cells. In this case, traditional methods encompass marking individual cells, just like putting colored stickers on the vehicles. However, this can be technically difficult and restricts researchers to specific periods.
Notably, GEMLI (Gene Expression Memory-based Lineage Inference), a cutting-edge tool for such purposes, was invented by Dr. Almut Eisele et al., which is capable of tracking cellular ancestry only through monitoring their gene expression patterns – the particular sets of genes that are active in a cell at any given moment. By introducing the concept in their recent Nature Communications article, novel approaches negate extra experiments, allowing easy and keen study of lineage.
What Determines GeMLI Success?
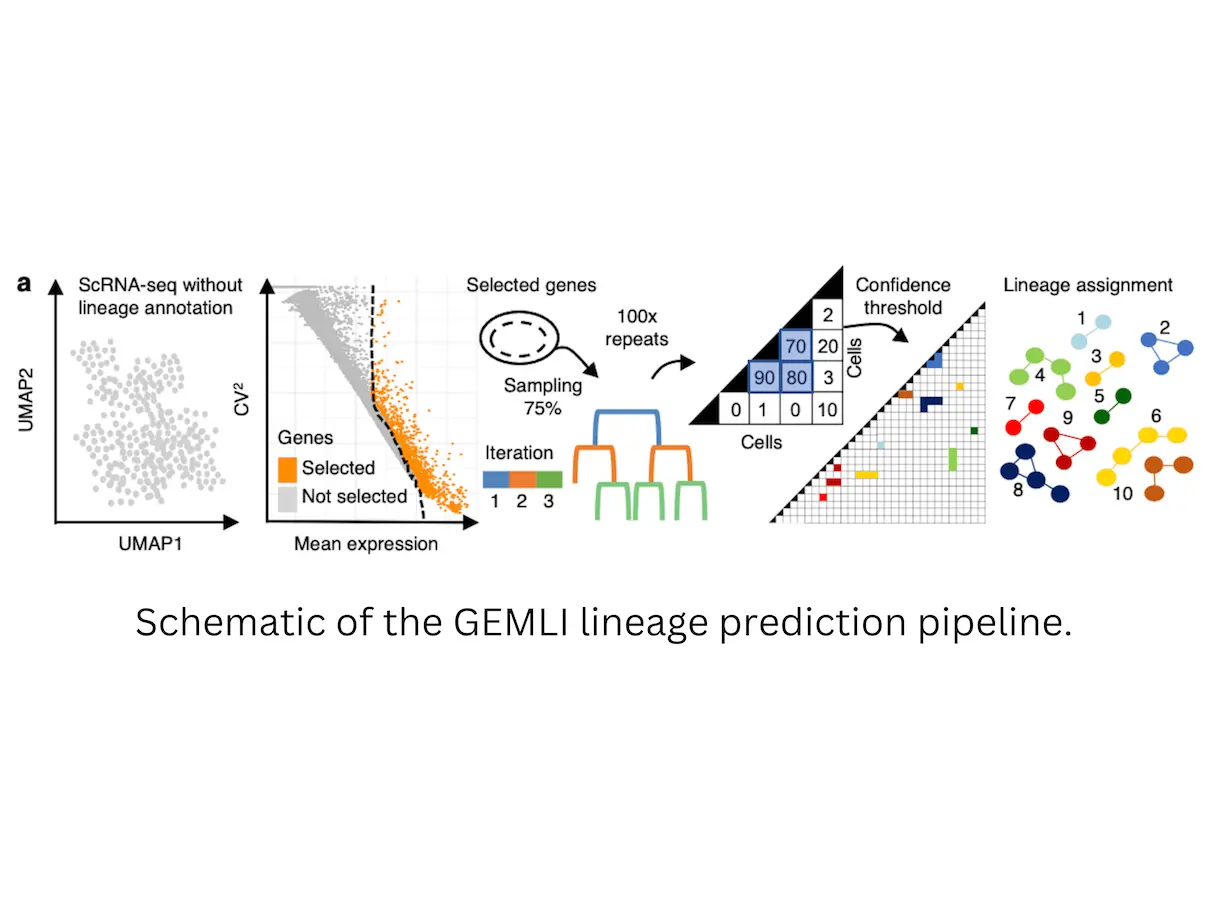
Ultimately, it all comes down to a very interesting thing called gene expression memory. Some genes are like reliable family storytellers that keep their expressions constant as cells divide and multiply. These “memory genes” contain information about the recent history of a cell, showing them as siblings, cousins, or other relatives.
GEMLI capitalizes on this by detecting genes with consistent expressions across several cells. This is where it becomes interactive! Visualize a scatterplot in which each point represents a cell, and its position reflects the activities of two memory genes. The clustering of cells suggests shared ancestors as reflected by similarity in gene expression profiles among these cells. By examining these clusters of memory genes across large data sets, GEMLI can reconstruct complex cell-family trees that shed light on lineage relationships between hundreds or thousands of cells.
Cellular Wonders Unearthed by GEMLI
The potentials for GEMLI are wide-reaching and greatly useful to researchers from diverse areas. Here are some examples:
- Understanding Development: Tracing stem cell lineages as they differentiate into specific tissues can be invaluable for understanding how organs and organisms form.
- Regeneration: It is the process through which new tissues are formed and damaged or injured ones are repaired. It can also help develop regenerative therapies in medicine by studying the lineage of cells involved in tissue repair.
- Cancer research: This kind of research can show how cancer cells arise from normal ones and secondly identify their vulnerabilities according to their lineage that might be used for therapeutic purposes.
Disease progression unfolded: In this case, GEMLI tracks immune cell lineages as well as disease-causing microbes, thereby showing how diseases spread inside the human body.
GEMLI has several other uses. Some of them include:
- Cell Fate Determination: Asymmetric divisions occur when two cells arise from a single parent and differentiate into two distinct cell types differing in structures. These decisions regarding fate during asymmetric division can be made with the aid of GEMLI, which analyzes memory gene expression patterns.
- Multi-cellular Structure Reconstruction: Suppose we take an example of complex tissue like skin; by evaluating individual cells from such tissue, it becomes possible for GEMLI to rebuild the original spatial organization relying on inferred lineage relationships of these cells.
Beyond the Basics: An In-Depth Look at GEMLI
The GEMLI world is full of awesome technicalities for those who want to dig. The study by Eisele et al. describes specific criteria used to identify memory genes and algorithms for lineage inference. This powerful tool can be implemented in various studies by researchers through the freely available GEMLI R package on GitHub.
For curious minds, let us look into some of the ongoing debates around GEMLI:
Limitations and Future Directions: However, it is necessary to acknowledge some limitations to GEMLI despite being a huge leap forward. The accuracy of lineage predictions depends on scRNA-seq data quality and the presence of informative memory genes. Consequently, future research may focus on refining the process of selecting memory genes as well as eventually incorporating other data sources, such as spatial information.
Ethical Considerations: As our ability to map cellular lineages improves, ethical considerations arise. Can we use lineage information to target or identify specific cell populations in therapies? Thus, open discussions among scientists, ethicists, and policymakers will be key in ensuring responsible use of this powerful technology.
Unveiling of the Cellular Tapestry
GEMLI can predict cell lineage from single-cell RNA sequencing (scRNA-seq) data. Gene expression memory is what GEMLI explores in its ability to analyze and study gene expression memory through the entire family tree of cells; it affords a new platform to explore research in various fields. Let us further intertwine the strands in this intriguing technology by looking at its impact on various scientific disciplines and the amazing cellular lineage studies.
A Boon for Biological Discovery:
- Developmental Biology: If you could turn back time and see a zygote change into an adult organism, wouldn’t that be amazing? Stem cells help to form different types of cells in organs and tissues as they develop during embryogenesis. This allows us to understand intricate choreography inside individual cells throughout the development process that may reveal reasons for congenital malformations or give direction toward regenerative treatment.
- Regeneration and Repair: The body has an incredible capacity to self-repair. For instance, GEMLI enables researchers to study the lineage of tissue-regenerating cells such as liver or skin derivatives. Developing strategies that could enhance the natural healing processes of a person or even create new ways of regenerative therapy can be possible through understanding their origination and differentiation.
- Cancer Research: It offers fresh insights by enabling researchers to follow cancer cells as they emerge from normal ones. This way, the origins of cancer cells can be discovered, and lineage-specific vulnerabilities may be found for better therapies.
- The immune system: The immune system is an elaborate network of cells that keeps our bodies healthy. In the case of T-cells and B-cells, GEMLI helps us keep track of their development as well as their lineage formation during the pathogen recognition and fighting process. Such a discovery paves the way for designing immunotherapies that are more specific in dealing with different diseases.
- Neuroscience: Billions of neurons connect, which provides us with a complex maze called the human brain. Using GEMLI would make it possible to trace back these neurons’ lineages, thus shedding light on how neuronal circuits are formed during development, thereby assisting in findings about neurodegenerative disorders like Alzheimer’s.
GEMLI Future: Challenges and Opportunities
As with any cutting-edge technology, there are certain hurdles and exciting prospects for GEMLI in the time to come:
- Bounds and Tidying: The precision of lineage predictions depends on the quality of scRNA-seq data and the presence of informative memory genes. In future studies, refining the process of selecting memory genes may be a better option, as other data sources like spatial information might be included.
- Ethical Considerations: The deeper scientists go into cellular lineages, the more ethical issues arise. Is it possible to use lineage information for the identification and targeting of special cell populations for therapeutic purposes? Responsible use is only achieved through open discussions between ethicists, scientists, and policymakers.
- Integration with Other Technologies: The future lies in integrating GEMLI with other state-of-the-art techniques. A more holistic understanding of cellular behavior within tissues can be obtained by combining lineage information with protein expression data or spatial transcriptomics.
- Open-source Collaboration: For example, the freely available GEMLI R package on GitHub provides an enabling environment for scientific collaboration. Ongoing development can be contributed to by researchers from around the globe.
Concluding Remarks: The Cellular Tapestry in Unveilment, One Lineage at a Time
GEMLI embodies the paradigm shift that dramatically increases our ability to study cell lineages. Examining memory of gene expression offers a powerful tool for unveiling an intricate tapestry of cellular relationships within our bodies. This technology is transformational and can be used to uncover developmental secrets, regeneration, disease progression, and numerous other biological processes. Further improving GEMLI will lead to even more revolutionary findings in the future when it is merged with other state-of-the-art technologies. The journey toward unraveling the cellular tapestry has just begun, and GEMLI provides us with a strong thread that helps us better understand the cellular world.
Article Source: Reference Paper | GEMLI is available as an R package on GitHub
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.