Cancer is a complex disease. It is brought on by the accumulation of several gene alterations. Understanding the patterns of these alterations, or mutational signatures, has been crucial to comprehending the genesis, prognosis, and available therapeutic options for cancer. However, there is a big obstacle in trying to extract these characteristics from the abundant genetic data. But the techniques used to analyze these fingerprints so far haven’t been flawless. This approach’s potential is limited by the difficulties associated with appropriately assigning weights to existing signatures and identifying new ones. This is where MuSiCal (Mutational Signature Calculator), a potent new analytical framework created by Harvard Medical School scientists that seems set to transform the industry, enters. By reanalyzing more than 2,700 cancer genomes, researchers provide an improved catalog of signatures and their assignments, discover nine indel signatures absent in the current catalog, resolve long-standing issues with the ambiguous ‘flat’ signatures, and give insights into signatures with unknown etiologies.
Introduction
Cancer is a complicated illness caused by the accumulation of mutations in several genes. Understanding the patterns in these changes, referred to as mutational signatures, provides a potent window into the inner workings of the illness.
Distinct signatures linked to particular cancer subtypes or even individual tumors enable:
- Precision diagnosis
- Prognostic insight
- Targeted therapies
Mutational signatures give vital clues about the role of the environment in the development of cancer. The identification of indications of carcinogen exposure, such as UV radiation or tobacco smoke, can help drive preventative and public health measures.
Analysis of mutational signatures has become a useful technique for identifying the mutational pathways behind somatic DNA alteration. Robust algorithms and a thorough grasp of mathematics are needed to enhance the accuracy and interpretability of mutational signature analysis and maximize its impact. Computational methods in the field have developed quickly in the past ten years. Popular tools such as SigProfilerExtractor, SignatureAnalyzer, signature.tools.lib, and others have achieved considerable success.
However, several methodological difficulties remain:
Signature assignment: The challenge of precisely determining the contributions of active signatures within a sample or dataset.
- Similar-patterned “flat” signatures are especially hard to differentiate.
- Mistaken assignment may result in inappropriate treatment decisions.
Signature discovery: It is challenging to compare results and uncover new signatures since current approaches yield inconsistent results.
- Due to algorithmic biases, a large number of found signatures may be variants of existing ones.
- This may obscure tissue-specific indicators and complicate our interpretation of their genesis.
Researchers introduce MuSiCal, a complete framework that allows robust and sensitive signature discovery and correct signature assignment. MuSiCal uses several novel techniques, such as minimum-volume NMF (mvNMF), likelihood-based sparse nonnegative least squares (NNLS), and a data-driven strategy for systematic parameter tuning and in silico validation, to address the issues mentioned above. Furthermore, by reanalyzing over 2,700 genomes from the Pan-Cancer Analysis of Whole Genomes (PCAWG) project, the study provides a better catalog for both signatures and their assignments to aid future investigations of mutational signatures in cancer. With nine additional ID signatures that are not included in the current COSMIC collection, the new ID signature library is more comprehensive. By resolving problems with ambiguous flat signatures, the derived signature assignments also shed light on signatures with unclear etiologies.
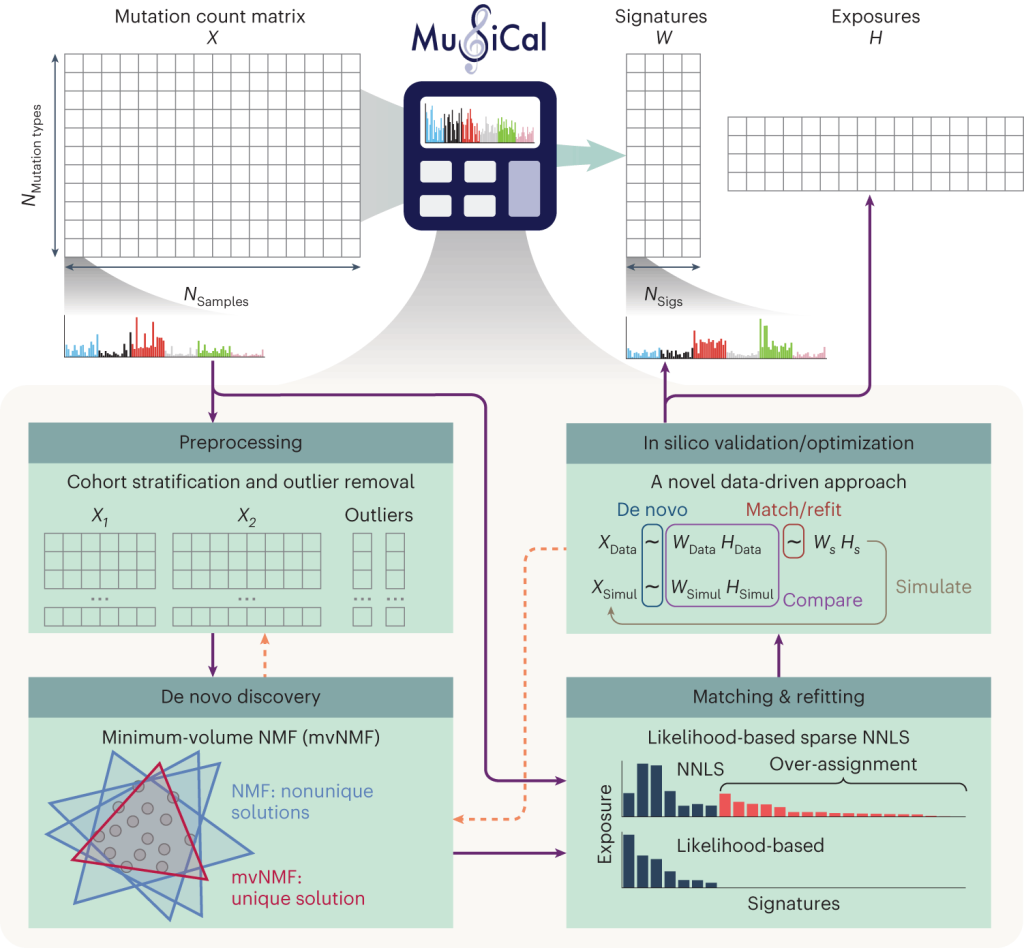
Image Source: https://doi.org/10.1038/s41588-024-01659-0
De novo mutational signature discovery with mvNMF
MuSiCal’s capacity to identify de novo signatures is one of its primary benefits. Unlike conventional approaches that rely on pre-established signatures, MuSiCal can detect wholly novel signatures directly from the data, regardless of prior knowledge. This might lead to groundbreaking discoveries since it allows researchers to uncover novel mutational pathways peculiar to specific cancer types or even individual tumors.
The core technology behind MuSiCal’s de novo discovery is minimum-volume Non-negative Matrix Factorization (mvNMF).mvNMF is guaranteed to recover the true underlying signatures under mild assumptions satisfied by tumor somatic mutations, whereas NMF requires much stronger assumptions that are often unrealistic. Researchers used NMF and mvNMF to construct synthetic datasets with Dirichlet-distributed exposures and tumor type-specific SBS signatures to make a more methodical comparison. When averaged across several signatures within the same tumor type, mvNMF reduces the cosine error by 67-98%, improving the accuracy of identified signatures when compared to NMF. The improvement is especially apparent in somewhat flat signatures. Due to interference from other similar signatures, even sparse signatures with nonzero weights in a small number of SBS categories may create serious issues for NMF; nevertheless, mvNMF remains unaffected by these distortions.
A Case Study: Reanalyzing the PCAWG Data
To showcase the real-world impact of MuSiCal, let’s take a closer look at its application in reanalyzing the data from The Cancer Genome Atlas (TCGA) Pan-Cancer Analysis of Whole Genomes (PCAWG) project. Furthermore, the approach made it easier to identify signatures specific to POLE-exo mutations and mismatch repair deficit (MMRD), offering important new understandings of these clinically significant mutational processes.
Researchers reanalyzed more than 2,700 cancer genomes from PCAWG for both SBS and ID signatures to derive a refined catalog of signatures and their assignments. Their key findings are as follows:
SBS signatures
- Independent de novo discovery was carried out for each type of tumor.
- Outlier elimination and L1 normalization were used as preprocessing techniques.
- Refitted exposures and matched de novo signatures to the COSMIC database.
- Further signals unique to MMRD/POLE-exo mutations were found.
ID signatures
- Because there were fewer IDs per sample, a combined analysis of many tumor types was conducted.
- Tumor types are categorized according to exposure matrix clustering.
- A new ID signature catalog was derived.
A Tool for the Future of Cancer Research
Musical represents a significant advancement in the field of mutational signature analysis. It has become routine in cancer genome analysis. MuSiCal outperforms state-of-the-art algorithms for both signature discovery and assignment. MuSiCal’s improved performance is further demonstrated by the improved consistency of MuSiCal-derived signature assignments with biological ground truth in real data when such ground truth is known, as is the case for homologous recombination deficiency-associated SBS3 and platinum-associated SBS31/35.
The number of whole genomes of cancer and other diseases continues to grow rapidly, especially through consortium projects such as Genomics England and the Hartwig Medical Foundation’s metastatic tumor project. By applying MuSiCal to these datasets, the collection of mutational signatures will be improved, and comparisons between signatures from different contexts—such as tumor types, metastatic status, and drugs received—will be facilitated.
Conclusion
Mutational signature analysis is a recently developed computer method for analyzing somatic mutations in the genome. Its use of cancer data has improved our knowledge of the mutational dynamics that drive carcinogenesis and shows its potential to guide prognosis and therapy decisions.
Researchers present the MuSiCal, a rigorous analytical framework with algorithms that solve major problems in the standard workflow. By reanalyzing over 2,700 cancer genomes, they provided an improved catalog of signatures and their assignments, discovered nine indel signatures that are not in the current catalog, resolved long-standing issues with ambiguous ‘flat’ signatures, and provided insights into signatures with unknown etiologies.
It is anticipated that MuSiCal and the updated catalog will be a step toward establishing best practices for mutational signature analysis.
Article source: Reference Paper | MuSiCal is implemented in Python and is available on GitHub
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.
















