For decades, tracing the lineage of individual cells has been a difficult and unreliable task. Existing methods like dye labeling and genetic barcoding have many limitations. To tackle these limitations and problems, a group of researchers from the California Institute of Technology, USA, developed “baseMEMOIR,” a multiplexed phylogenetic recording system that enables detailed recording of lineage relationships over time in a manner compatible with the recovery of spatial position and gene expression patterns. It is a revolutionary solution borrowing the power of base editing, a precise genetic engineering technique to turn cells’ DNA into a record keeper.
Introduction
Lineages and historical states of individual cells within a tissue or organism refer to an important concept in biology called cell lineage tracing. It is essential for understanding the family tree of cells but on a much smaller level. Lineages are the “family” of the individual cell, and historical states are the different “stages” that it has gone through in its life. Knowing these two can answer many questions, like how different cells emerge and organize themselves into different complex structures, cell fate, or their response to various stimuli. Throughout this journey of tracing the cell lineage, many tools and methods came into being but were not up to the mark. Existing methods, like lineage tracing dye, Fluorescent protein markers, and cell lineage barcoding with CRISPR/Cas9, have limitations like limited resolution, short time scale disruption of natural tissue architecture, and many more.
This is where BaseMEMOIR came to the rescue. It was carefully designed and tested by a group of researchers to tackle the maximum limitations of preexisting tools and methods. BaseMEMOIR turns cells’ DNA into record keepers by using the power of a precise genetic engineering technique that is base editing. Specific DNA sequences are designed within the genome by engineers that can be selectively altered by the base editor. These sequences act as tiny switches, flipping between three distinct states detectable under microscopy, where each state signifies a different step in the cell’s history, creating a visual representation of its past.
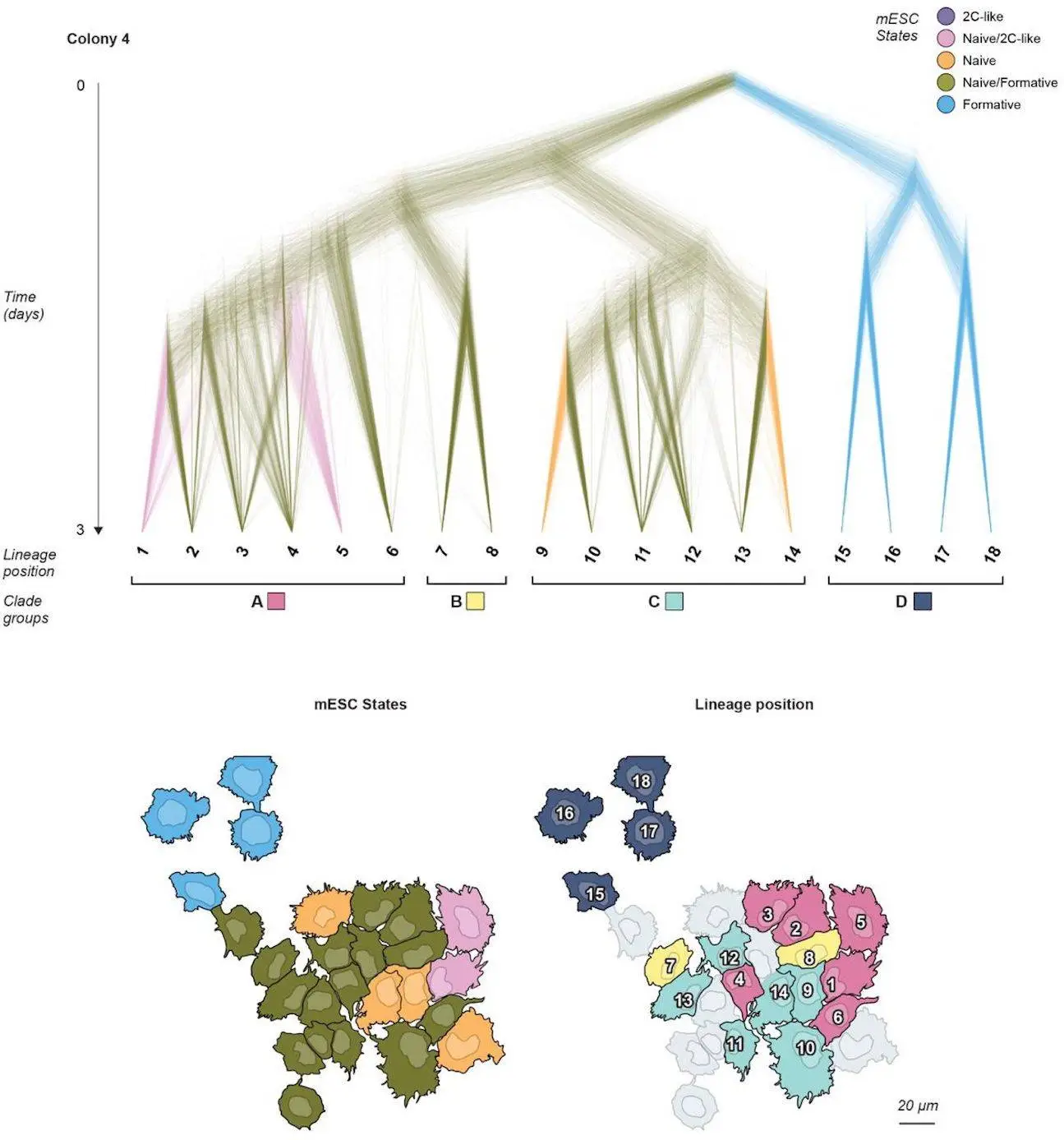
With the help of BaseMEMOIR, scientists can now encode up to 792 bits of information per cell, which is a 50-fold increase over previous methods and allows for detailed reconstruction of cell lineage trees. It also maintains the spatial context of the cell and allows the recovery of gene expression patterns. Scientists applied BaseMEMOIR to stem cells to test it. They were able to reconstruct a lineage tree for seven colonies with up to 4-7 generations per colony, along with its influence on Wnt (through CHIR) on state dynamics.
Limitations of the existing tools :
1. Fluorescent protein markers: Lose signal through cell divisions, limited number of colors, phototoxicity potential.
2. Lineage tracing dyes: Short timescale, limited information about cell history, can be toxic to cells.
3. Retroviral lineage tagging: Invasive, irreversible, not suitable for all cell types, prone to insertional mutagenesis.
4. Single-cell RNA sequencing (scRNA-seq): Static snapshot, expensive, doesn’t capture cell dynamics or lineage relationships.
5. Cell lineage barcoding with CRISPR/Cas9: Complex to design and implement invasive, potential off-target effects.
Scientists used various methods for constructing BaseMEMOIR to ensure its perfect, limitation-free functioning. Major steps which were involved were:
- Plasmid construction and cell line engineering
- Next-generation sequencing (NGS)
- Zombie-FISH preparation and imaging
- Image processing and barcode classification
- Cell type analysis
- Lineage reconstruction and Bayesian modeling
- Lineage motif analysis (LMA)
Based on the above steps, the researchers drew various conclusions and determined key components and strategies of BaseMEMOIR.
Key components of BaseMEMOIR system:
- Base editor: BaseMEMOIR system uses a lineage tracing method, which is the combination of a base editor (ABE), a DNA barcode array, and sequential fluorescence in situ hybridization (FISH).
- Dynamic barcodes: BaseMEMOIR generates diverse editing outcomes over time with the help of Six target sites for ABE editing
- Static barcodes: BaseMEMOIR uses 40,000 unique sequences for tracking individual cells. Sixty-six unique statically barcoded target arrays collectively provide 792 bits of editable information in the baseMEM-01 cell line.
- Zombie-FISH: In BaseMEMOIR for sensitive detection, amplify barcode signals. The zombie readout system allows 4-way probe competition while maintaining compatibility with the FISH-based readout of endogenous genes.
- Machine learning: For accurate barcode state identification, BaseMEMOIR uses an SVM classifier.
- Bayesian modeling: BaseMEMOIR uses the BEAST2 framework to reconstruct lineage trees and estimate cell motility by adding a new mutation model and taking advantage of its phylogeographical and discrete trait models.
- Lineage motif analysis (LMA): Common cell fate patterns identification.
Advantages over Existing Lineage Tracing Methods:
- Higher resolution: BaseMEMOIR combines dynamic barcodes with static barcodes, offering detailed information about individual cell history compared to existing tools.
- Longer timescale: Using dynamic barcodes in BaseMEMOIR enables tracking across multiple cell divisions, overcoming limitations of methods reliant on short-term markers.
- Single-cell level analysis: BaseMEMOIR provides data on individual cells, which reveals heterogeneity and rare events, unlike bulk population assays.
- Quantitative lineage reconstruction: BaseMEMOIR uses Bayesian modeling, which allows rigorous statistical inference by enhancing data interpretation of cell state transitions and lineage relationships.
- Identification of common cell fate: Using lineage motif analysis tools like LMA BaseMEMOIR enables the discovery of recurring patterns in cell lineages, which provides insights into developmental processes.
- Flexibility: Customization of BaseMEMOIR is possible for different cell types and tissues by targeting specific genes and designing relevant barcodes.
- Scalability: Being adaptable to large datasets, the BaseMEMOIR system can be potentially applied to analyze complex tissues and organs.
Though BaseMEMOIR is a very powerful tool, it does have some limitations. For example, it does not directly probe the states of cells at earlier time points, and it cannot directly detect earlier states that do not appear in the endpoint measurement. Analyzing systems at multiple time points could help to avoid missing transient states. As it is a relatively new tool, these issues may be solved by further technical improvements to barcode imaging.
Breaking through the limitations of traditional methods and passing with flying colors, BaseMEMOIR has proved to be a game changer. It will be a great help for scientists in their novel findings and research. It will provide a useful tool for understanding development, diseases, and other biological processes that involve dynamic changes in cell lineage. It opens a new door for studying cell history at unpredicted scale and resolution.
Future applications of BaseMEMOIR may include:
- Stem cell differentiation and early mouse embryogenesis exploration.
- Diverse developmental and physiological processes adaptation.
- Spatial cell atlases with lineage information augmentation.
- Role of lineage, signaling, and differentiation in disease progression investigation.
Conclusion
It was the long-standing dream in biology to image a tissue or organism and visualize not only its current state but also its history. Previously, many tools and methods have tried to do so but failed miserably. This is where BaseMEMOIR came to the rescue; it was developed by a group of scientists after great difficulties. It provides much larger memory sizes and allows for deeper, more accurate lineage tree reconstruction while preserving spatial structure. To achieve this, it introduces several key innovations like the use of base editors to introduce stochastic but precise edits at dense target arrays, the use of dinucleotide editable target sites by incorporating 66 unique statically barcoded target arrays to expand the amount of memory accessible in single cells, etc.
To demonstrate BaseMEMOIR capabilities, researchers applied it to stem cells undergoing interconversion among transcriptional states, which resulted in the reconstruction of lineage trees for seven colonies totaling 197 cells, with as many as 4-7 cell generations per colony. BaseMEMOIR, being a new tool, contains some limitations, but those issues can be addressed with improvement in technology. Its future is very bright and could be readily adaptable to diverse developmental and physiological processes.
Overall, it is a fantastic tool for studying lineage and cell state dynamics in multicellular systems.
Article Source: Reference Paper
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.