A new tool from UC Berkeley can predict mutations that can improve the way how RNA works. By leveraging hyperthermophilic RNAs, the researchers identified mutations conferring increased thermostability to E. coli ribosomes, providing a foundation to understand RNA sequence-structure-function relationships.
Introduction
Do you remember what DNA is? It’s that molecule that contains all the information necessary for life, isn’t it? Well, its less-known cousin – RNA, has a starring role in translating this blueprint into action. In protein production as well as gene regulation processes, RNA is an adaptable and powerful player. Nonetheless, forecasting how structure influences function in RNA has been one of the toughest jobs.
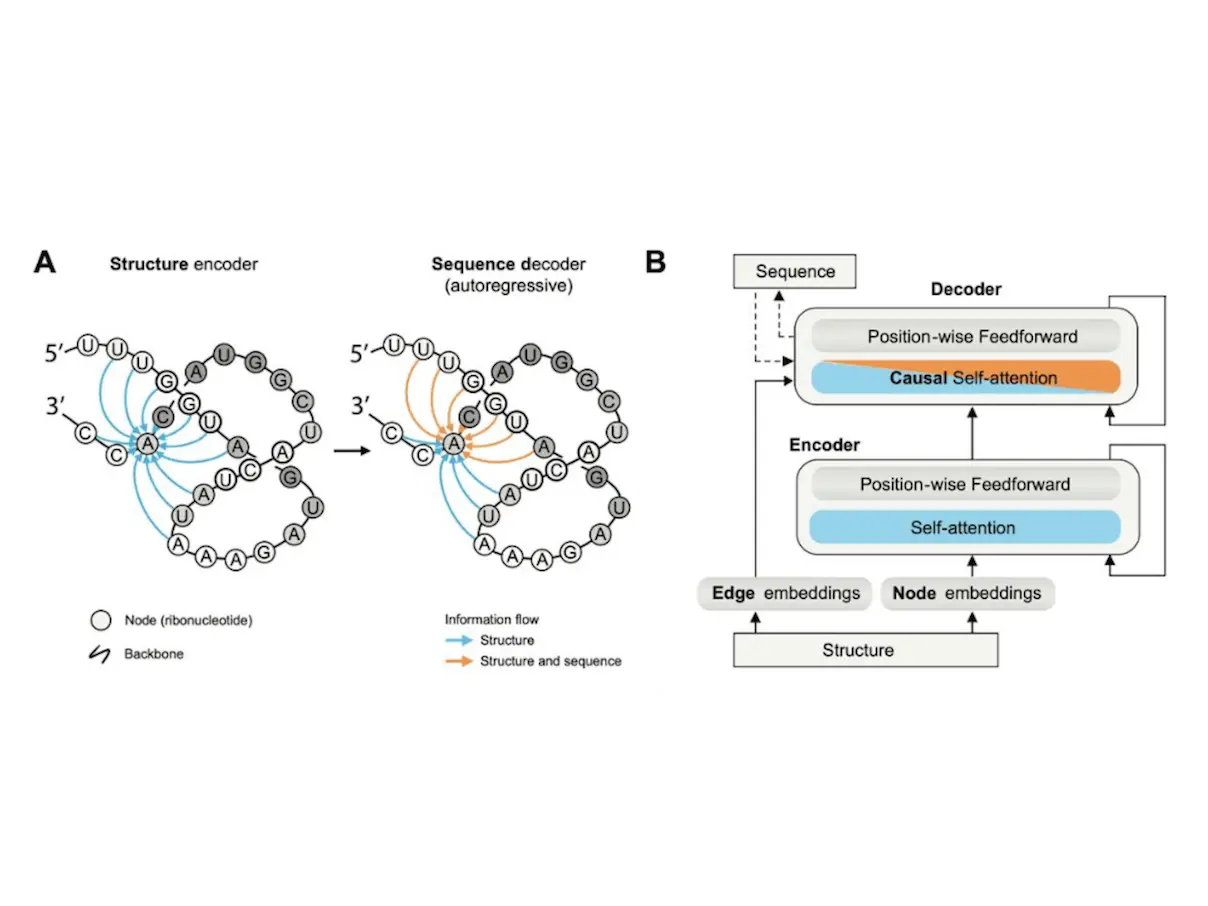
RNA language models have revolutionized everything. This isn’t just your ordinary spellchecker for genetic code but a profound AI tool that not only identifies possible alterations in RNA sequences but also predicts their influences on the functionality of this molecule. Hold on, Science lovers! A paper by Shulgina et al. has landed, and it’s going to change everything we thought we knew about the future of RNA research.
The Tricky Prediction of Function: The Conundrum of RNA Structure
While DNA structures contain a double helix with well-defined shapes, RNAs take complex forms when folded into 3D shapes. Such shapes are vital for performing RNA functions. Slight modifications within chains of adenine (A), uracil (U), guanine (G), or cytosine (C) can turn around this 3D shape, thus making it useless or toxic.
What’s the problem? Traditionally, predicting RNA structure from its sequence has been a formidable task due to the paucity of good data and the complicated manner by which RNA folds. This constricted our knowledge about how mutations and modifications affect RNAs.
GARNET: An RNA Functional Rosetta Stone
To address this data limitation, the researchers of this pioneering research work have developed GARNET (Gtdb Acquired RNa with Environmental Temperatures), which is an exclusive database. A kind of Rosetta Stone for RNA function, GARNET unleashes an abundance of information. It doesn’t just store RNA sequences; it links them to the organisms they come from and even factors in the organism’s optimal growth temperature.
This seemingly insignificant detail – temperature – has a minefield of secrets hidden within it. Organisms that live well in extremely high temperatures (hyperthermophiles) have RNA molecules that are adapted to withstand these conditions. Through a comparison between these “heat-resistant RNAs” and their cold-loving counterparts, scientists can identify sequence features contributing to different temperature stability/functionality relationships for RNAs.
Building an AI Model for RNA: The Power of Language
The researchers went a step further, using GARNET to provide a rich tapestry of RNA data. They utilized deep learning, a form of artificial intelligence, and created RNA’s unique “language model.” Imagine a super-charged spell-check on acid! This AI model can not only detect mistakes in the sequence of RNAs but also predict how changes in sequence (mutation) might affect the 3D shape and, hence, its function.
Here is the beauty of this approach. Traditional ways to study RNA function include laborious trial-and-error methods involving making mutations and observing their effects. However, through the use of the RNA language model, scientists have a powerful predictive tool at their disposal.
Engineering Heat-Resistant Ribosomes: The Proof is in the Pudding
They built not just some expensive model; they took it out into the real world and tested it with the real stuff. They focused on ribosomal RNA (rRNA), an essential component of ribosomes – cellular machinery responsible for protein synthesis. Analyzing heat-resistant rRNAs from hyperthermophiles in GARNET helped to identify specific mutations that could improve the thermostability (heat resistance) of E.coli ribosomes as predicted by the model.
Guess what? The model predictions were right! Indeed, the introduction of predicted mutations into E. coli rRNA made the ribosomes more resistant to heat. This successful experiment shows that RNA language models are powerful and can accurately predict the functional consequences of mutations.
The Future of RNA Research: Infinite Possibilities
The impact of this research is truly transformative. Think about being able to:
Design RNAs having different functionalities like;
- Next-generation drugs: We could build RNAs that affect certain disease-causing molecules or even deliver gene therapies exactly where they are needed.
- Biosensors: Artificially created RNAs can be bio-detectors that sense specific molecules or environmental changes.
- Advanced biomaterials: Tailor-made RNAs could be used to make unique materials with novel properties for a range of applications.
Indeed, nothing is impossible. With RNA language models, we not only see how RNA works, but we can also design them as architects by manipulating them to achieve particular goals.
Beyond the Hype: Troubles and Considerations
In addition, there are still a few challenges that need to be met with this research heralding a new era of RNA manipulation. The accuracy of these models depends mostly on data in GARNET that is both abundant and of good quality. Such sophistication and reliability will increase as more functional data and RNA sequences are incorporated. Further study is needed to verify how well laboratory findings translate into real-life situations using living organisms.
Ethical Considerations
However, the most important question here is how ethical it is to have the ability to manipulate RNA function. Just like any other powerful technology, the use of RNA language models should be approached with caution regarding possible misuse. This calls for responsible research practices concerning off-target effects, unforeseen consequences, and potentially dangerous RNA molecules under clear ethical guidance.
Open science is crucial in ensuring transparency and encouraging collaboration through the sharing of research data and methodologies. This enables critical scrutinizing, refining, or building upon these preliminary results by the scientific community. Also, open-source RNA language models would make this tool accessible by democratizing it, thus allowing researchers from every corner of the globe to participate in improving our understanding of RNA.
Beyond the Hype: Extending the Scope of RNA Language Models
There is much more to the impact of RNA language models than engineering heat-resistant ribosomes. Here are a few other areas with huge potential for this technology;
- Understanding RNA-related ailments: Many diseases, such as brain disorders and some cancers, are a result of misfiring RNA molecules. The use of these models can help identify the exact mutations that cause these diseases, leading to the development of precise treatments.
- Optimizing RNA therapeutics: In recent years, there has been rapid growth in using RNAs to treat a plethora of illnesses. Using this model, one could rapidly design and refine RNA therapeutics by predicting their efficacy as well as potential side effects.
- Develop new tools based on RNA: Various applications, including gene editing and targeted drug delivery, have been touted for making use of RNA among researchers. When designing such tools that work optimally with limited off-target effects, it’s essential to consider the role of rRNA language models.
Conclusion: New Dawn of RNA Research
The advent of RNA language models represents a crucial juncture in our comprehension and manipulation of RNA. This mighty AI tool paves the way for a new era in RNA research where scientists can predict function, engineer novel functionalities, and possibly change medicine, biotechnology, and more.
Nevertheless, this research has only just begun. Consequently, further improvement should be made to open data sharing and responsible research practices so that this revolutionary technology is applied safely and ethically.
One thing is certain as we plunge into the world of RNA – the future looks promising. With the guidance provided by RNA language models, we can unlock its vast potential as well as redefine what it means to be involved in RNA research.
Article Source: Reference Paper | The Code used in this study is publicly accessible on GitHub | Associated data and the GARNET Database are available at https://tinyurl.com/5abszup9.
Important Note: bioRiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.