The complex processes that regulate biological systems have fascinated scientists for thousands of years. However, elucidating these complex issues sometimes requires thinking outside the box using traditional experimental methods. Here, computer modeling can help scientists gain valuable insights by replicating biological processes in silico. However, a major obstacle can be the amount of technical knowledge required to build and use these models. To solve this problem, researchers from the Medical College of Wisconsin developed BioModME, a simple online tool designed specifically for developing and analyzing models of biological systems. This blog explores aspects of BioModME, focusing on the benefits it offers researchers and how it can radically change the way biological modeling is done.
Bridging the Gap between Bench and Code
The study of biological systems has always depended heavily on experiments. Although this approach is extremely useful, it can be expensive, time-consuming, and limited to deal with the ever-increasing complexity of biological networks. Due to a large number of interacting components, it is difficult to analyze these networks individually experimentally. In addition, the roles of certain elements can be unclear, making it difficult to understand how the network works.
Computer modeling offers an attractive alternative. Computer simulations of biological systems allow scientists to generate testable hypotheses for further study and to predict the likely behavior of a system under different conditions. However, most biologists find it difficult to achieve the technical expertise needed to build, implement, and solve these models. BioModME overcomes this drawback by providing an easy-to-use graphical user interface (GUI) that does not require prior coding skills.
Unveiling BioModME’s User-Friendly Interface
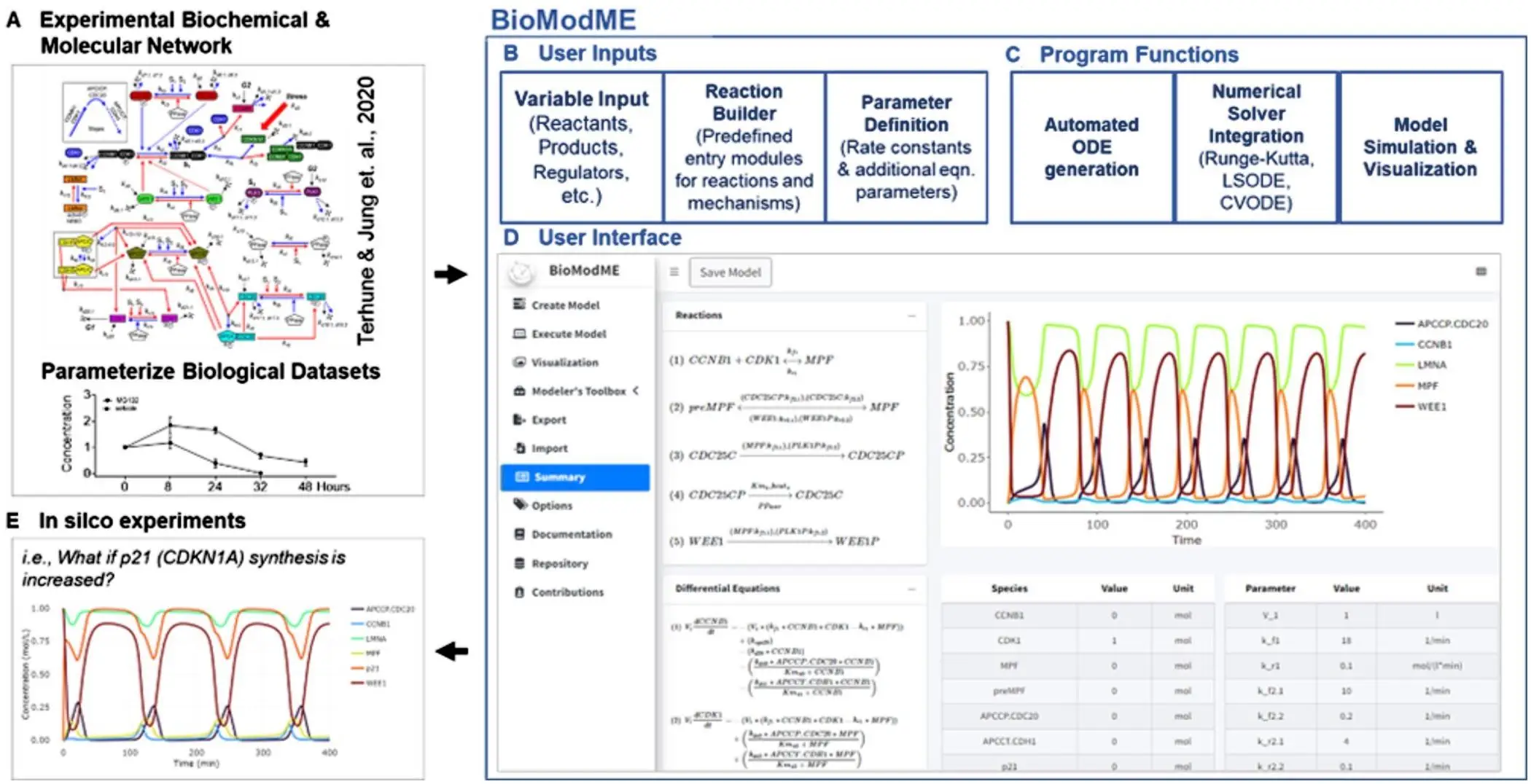
An open-source R web application called BioModME was created using the shiny package. This offers several significant advantages to consumers:
Ease of use: BioModME does not require the installation of applications because it works entirely through a web browser.
User-friendly interface: Users can create response networks, build models, and easily evaluate biological system performance thanks to a friendly graphical interface.
No Coding Required: Unlike traditional modeling methods, BioModME allows researchers to create models without writing a single line of code.
Building Robust Models with BioModME
BioModME offers a wide range of features to help create reliable biological models:
Multi-domain modeling: Researchers can create models of complex biological systems with multiple compartments or domains.
Customizable feedback and rules: better describe the nuances of reality and biological processes, BioModME allows users to add control mechanisms and create their response functions.
Unit management: The application has an integrated unit converter that handles multiple units of measurement to ensure model consistency.
Unveiling the Hidden: Model Simulation and Analysis
BioModME is not just about building models. It helps researchers evaluate and justify their findings.
Automatic ODE/DAE generation: BioModME eliminates the need for labor-intensive mathematical managers by converting a user-specified model into appropriate mathematical equations (ODE or DAE).
Simulation and visualization: Simulating model behavior and results presented as excellent visualizations, the application provides researchers with important insights into system dynamics. BioModME includes methods to estimate model parameters from experimental data, leading to more accurate simulations.
Model Comparison Suite: By simultaneously running simulations with multiple parameter values, researchers can evaluate model performance under different conditions.
Sharing is Caring: Data Exchange and Export Options
BioModME encourages research collaboration and data exchange by providing comprehensive export and data exchange functions.
SBML import/export: The tool allows the import of models in the widely used System Biology Markup Language (SBML) to share software systems.
Rich export options: BioModME offers several export options for model data, including CSV, XLSX, PDF tables, R or MATLAB code scripts, and mathematical equations in text and MathML formats.
Looking Ahead: The Future of BioModME
BioModME developers are constantly working to improve its features and functionality. Here’s a preview of what’s to come:
Sensitivity Analysis Tools: A set of tools to examine changes in model output as the parameters change.
Graphic UI: Create a visual UI using a JavaScript template that lets you drag and drop elements to visualize model paths.
BioModME: A Boon for Biological Research
BioModME stands out from other modeling platforms with a unique combination of features. All in all, BioModME is revolutionary in the biological modeling space. BioModME is free and open source, enabling it to be used by a wider scientific community. Regardless of coding background, researchers can create and evaluate complex biological systems models. It is a user-friendly and accessible online tool. A simple user interface eliminates the need for coding skills, offering versatile tools to build, model, analyze, and share biological systems. It does not store user patterns or session data on the host servers because of its strict user privacy policies. Users can securely save templates locally and share them with others.
Conclusion
BioModME provides a robust web application tool that helps researchers develop mechanistic computational models of complex biological systems containing multiple domains or compartments without requiring complex mathematical inference or knowledge of coding and solving mathematical equations in a programming language. Shiny’s user interface enables a guided model creation process that can be used to create robust model simulations and high-quality graphics.
The extensive capabilities of BioModME, including parameter estimation and multi-domain modeling, are useful for researchers in many biological fields. BioModME is free, open source, and web-based, making it easily accessible and encouraging collaboration and information exchange. BioModME expects further improvements, including tools for sensitivity analysis and graphical model generation. It paves the way for scientists with diverse coding experience to decipher how biological systems work and make breakthrough achievements in drug discovery, medicine, and other fields. BioModME makes biological modeling more accessible, enabling us to more quickly understand the complex dance of life.
Article Source: Reference Paper | BioModME can be accessed at Website | Source code is freely available for download at GitHub
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.