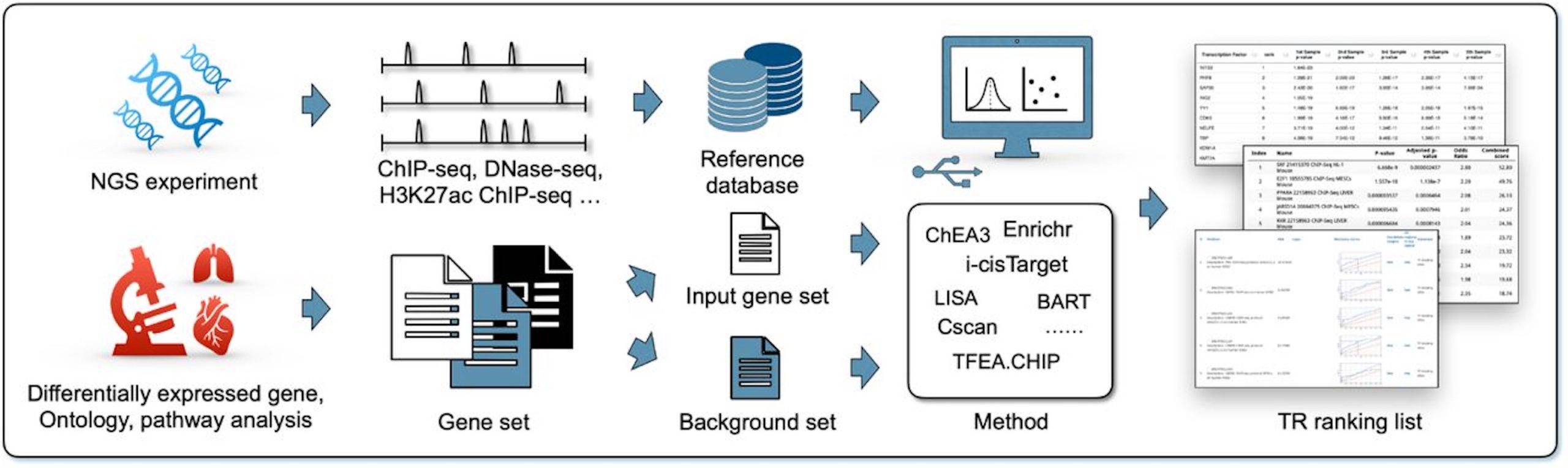
This article delves into a recent study undertaken by a team of researchers that examined multiple computer algorithms for transcriptional regulator prediction using Next-Generation Sequencing (NGS) data. Grasping biological processes and developing personalized medications necessitates a deep understanding of gene control. Transcriptional regulators (TRs) control the expression of certain genes, and NGS technology has enabled the discovery of these key players. This has opened up fascinating new study directions for scientists. It is hard to navigate the wide range of NGS-based approaches for TR prediction without some risk, though.
Introduction
It was a consequence of pioneering research and subsequent investigations that evolved the principles of gene transcriptional regulation in the 1960s. Researchers also revealed the crucial role of transcriptional regulators in controlling the transcription of hundreds of thousands of genes. They are proteins encoded by genes that can control transcription in a manner that directly influences gene expression levels. Also necessary for a wide range of cellular functions, including cell division, growth, morphogenesis, and death, determining TRs is essential for biological and medical research. Finding biomarkers and potential treatment targets can be aided by researching transcriptional regulators, as their data is linked to malfunctions and several diseases. It takes a lot of computational and experimental work to determine the relationships between TRs and their targets. Some sources like DEG analysis, gene ontology, and pathway enrichment analysis influence a user-defined gene collection, predicting them accurately.
The researchers concentrated on computational techniques, like epigenomic methods like ChIP-seq, ATAC-seq, and DNase-seq, which uses next-generation sequencing (NGS) technologies to identify TRs that regulate a certain gene set particularly. Unlike motif-based methods, NGS data may evaluate binding sites directly, which identify possible sites using computationally produced patterns.
Researchers begin this by:
- Firstly, they presented a comprehensive overview of the various NGS-based computational approaches, highlighting their similarities and differences.
- Secondly, providing a benchmark assessment using an extensive range of gene sets from TR perturbation research.
- Thirdly, they diligently examined the primary components of the eleven techniques, covering their sensitivity, coverage, accuracy, and usability.
They also checked their inadequacies and potential areas for future development. Then these methods were split into three categories:
- Library-based methods: These techniques use pre-compiled libraries of known regulatory interactions to identify potential regulators for your gene of interest.
- Regionally-based methods: These scan the regions of your gene’s DNA sequence for known regulatory patterns that may bind to certain regulators.
- Task-oriented strategies: These predict regulators based on many NGS-derived parameters using machine learning algorithms that have been trained on large datasets.
The scientists recognized the drawbacks of current methods and saw many possibilities for improvement. For example, the a need for user-friendly interfaces or to enhance predictions by merging data sources other than NGS and incorporating existing biological knowledge. They also highlighted the need for user-friendly interfaces and interpretable results in making these technologies accessible to a broader scientific audience.
Library-based methods
- Transform target genes into TR binding sites.
- Two subcategories are present:
- Window-based: Designate genes as targets using a predetermined window size (RegulatorTrail, ENCODE ChIP-seq significance tool, and Cscan).
- Window-free: Proximity decides which targets are allocated. (CHEA3, TFEA.ChIP, Enrichr).
Restrictions:
- Sharp cutoffs reduce feeling.
- Overlooking non-functional binding sites and three-dimensional chromatin structure.
- Different versions of enrichment analysis include gene set size bias, statistics, and FDR control.
Region-based methods
- Concentrate on paying attention to the general alignment of the regulatory elements and TR binding sites.
- There are two divisions:
- Nearest-elements-centric: Pay attention to the neighboring places and promoters. (ChIP-Atlas, MAGIC, i-cisTarget)
- Include distal elements: Include H3K27ac-specific distal regulatory components based on ChIP-seq results. (Lisa on BART)
Restrictions:
- Peak signal levels are not as reliable as functional indicators.
- Inadequate recording of distant CRE activity and its complex interactions.
- Unequal emphasis on research leads to distorted rankings based on data accessibility.
Key Findings
Accuracy, coverage, usability, and sensitivity are the four main parameters that researchers use to assess NGS-based techniques for TR identification. A systematic benchmark of thirteen methods by using 570 TR perturbation-derived gene sets, including accuracy, usability, coverage, and sensitivity.
- Top-performing methods: Remarkably, not a single one performs better than the others across the board. Lisa, ChIP-Atlas, and BART were the methods with the most reliable and accurate predictions.
- NGS vs. motif-based methods: NGS-based techniques perform better than motif-based techniques, while methods with large databases and region-centric approaches perform better.
- Limitations on accuracy: A rigorous interpretation and validation are crucial since even the best methods only rank around 10% of perturbed TRs in the top ten.
- Combining techniques: It was discovered by researchers that the use of a variety of tactics often yields better results than relying alone on one. Lisa and TFEA collaborated with ChIP, ChIP-Atlas, and BART, often providing the best performance.
- Coverage matters: Scientists also discovered that with more NGS data coverage and more recent publication dates, the performance of methods is higher.
- Obstacles & restrictions: The use of severe cutoffs and an unequal research focus may influence rankings. Researchers gave us a detailed walk-through along with the practical concerns and limitations.
- Usability: Enrichr, Lisa, ChIP-Atlas, and ChEA3 are well-maintained and user-friendly databases.
The Future Ahead
Even while researchers have proved NGS-based methods useful for finding TRs, they still have their limitations. To fully utilize their potential and get beyond any remaining limitations, more research is an absolute necessity. Complex projections are expected with time as long as data collection, resource integration, and algorithm development continue to advance and become more precise. Researchers should critically evaluate a mixture of different techniques with other strategies and appreciate their advantages and disadvantages to get a full understanding of gene regulation. The road ahead is difficult, but the fruit it will bear is unimaginable.
Conclusion
Identification of transcriptional regulators is of extreme significance in many biological applications, including but not limited to comprehending biological development mechanisms, identifying key disease genes, and predicting therapeutic targets. TRs should consider integrating single-cell data to improve their accuracy and resolution. In addition, with the rise of multi-omics experimental approaches, the availability of multi-omics data is no longer restricted to one or two types, and there is a growing need for novel multi-omics approaches that can leverage the full range of available data. Although pre-existing methods have been shown to perform well in their publication, a unified and systematic evaluation has not been conducted yet.
Several computational methods based on NGS data have been developed in the past decade, but no systematic evaluation of these has been offered. A group of researchers classified these computational methods into three categories on the basis of shared characteristics, namely library-based, region-based, and task-based methods; they further performed benchmark studies to assess the accuracy, sensitivity, coverage, and application of NGS-based methods with molecular experimental datasets. The findings show that BART, ChIP-Atlas, and Lisa have relatively better performance, offering competitive alternatives because of their reliable and accurate predictions. Despite their strength, the researchers also addressed the limitations of NGS-based methods and explored potential directions for further improvement in resource integration, data collection, and algorithm development, which might lead to even more accurate and nuanced projections.
It is expected that more multi-omics-based methods will be developed in the coming decade, which will undoubtedly lead to further innovation and discovery in this field.
Article source: Reference Paper | Commands and scripts to run each method is available on GitHub
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.