Unsung heroes of our cells are RNAs. These molecules are responsible for many biological processes, from carrying hereditary material to controlling protein production. However, imagine being able to change the functions of RNAs for medical purposes or scientific studies. In their latest research, scientists from the Massachusetts Institute of Technology and Broad Institute of MIT and Harvard, USA, introduce RNAFlow – an innovative approach for RNA sequence-structure design using inverse folding-based flow matching.
Creating the design of RNA molecules was traditionally a very long and wearying process. At times, existing methods would only focus on the sequence or structure of RNA rather than the intricate interplay between them, and they depended much on trial-and-error approaches. RNAFlow provides a novel way to design RNA molecules that incorporates both structure and sequence simultaneously, thereby providing an opportunity for the rapid development of new RNAs with desired properties. It relieves such shortcomings by utilizing the novel manner of using the structure prediction capabilities of RosettaFold2NA in RNAFlow
Power Generated by Flow Matching
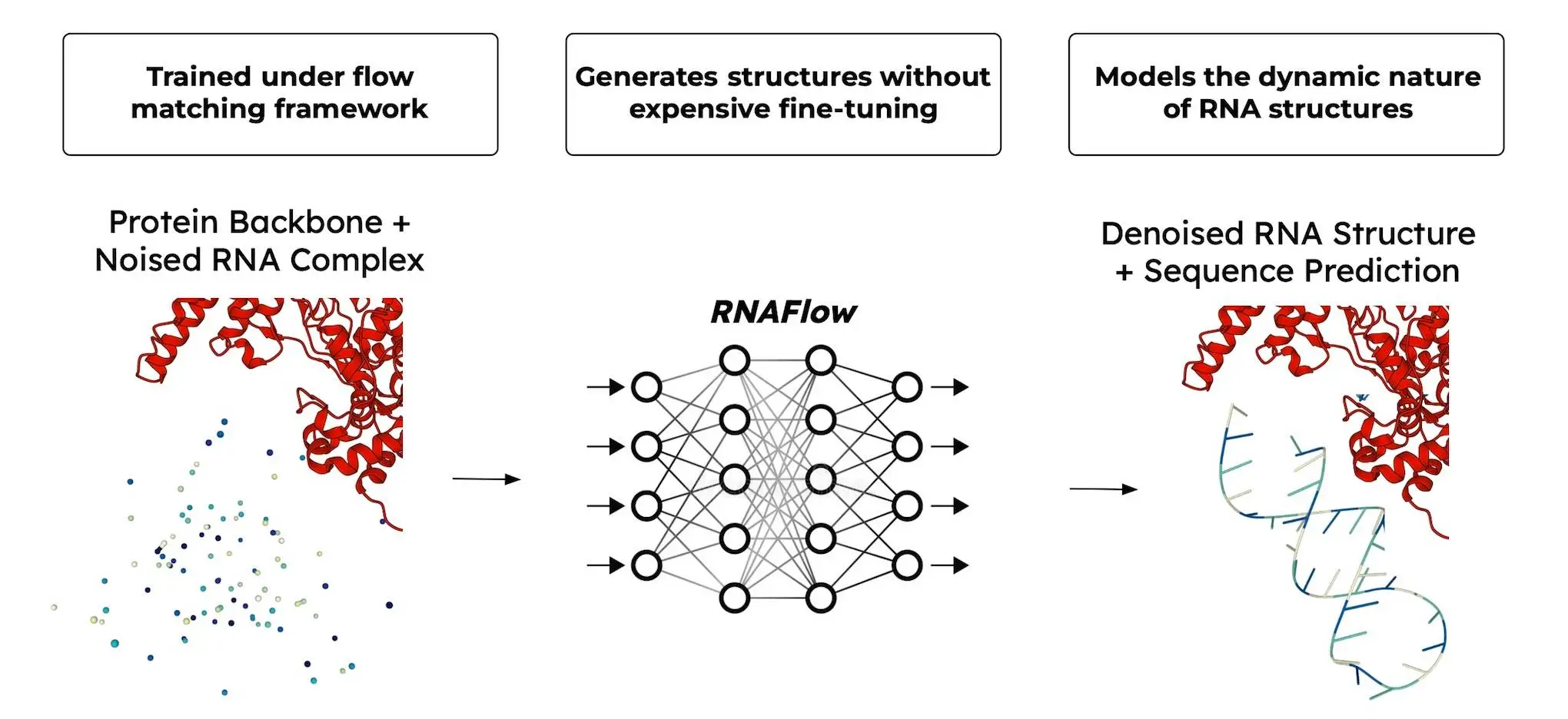
RNAFlow is a flow matching approach that resembles a river, which flows from an untidy, unorganized origin to a clear-cut destination. RNAFlow starts with a noisy version of the desired RNA structure and sequence. Afterward, the structure and sequence are iteratively refined until they resemble a perfect match.
Below is an outline of the key components of this iterative process:
- Noise-to-Seq: This section acts like a sanitation crew that converts noisy structures into clear and accurate ones (predicted by RosettaFold2NA).
- Traj-to-Seq: This step acknowledges that RNA structures are not fixed but can be represented by multiple conformations. By including these conformations, Traj-to-Seq facilitates designing RNAs with greater flexibility.
- Output Rescoring: All designs for generated RNA are not equivalent. In this aspect, it serves as a quality control mechanism for choosing the best candidates based on their predicted recovery rate.
The Backbone of RNAFlow is RosettaFold2NA, it is a modern protein structure prediction method. But here’s the exciting twist: it is not only capable of predicting proteins! Notably, RosettaFold2NA can predict 3D structures of RNA molecules, which is very important in RNA design. This property makes RosettaFold2NA the basis on which RNAFlow thrives.
Success on Multiple Fronts
The researchers carried out a variety of tests to determine whether RNAFlow is indeed helpful. They took into consideration the following points:
- Structure Generation Accuracy: How effective is RNAFlow in predicting 3D structure for an RNA molecule compared to other methods?
- Sequence Generation Accuracy: How well does RNAFlow generate an RNA sequence that folds into the target structure?
The results were impressive. In all cases, the structures and sequences generated by RNAFlow closely matched the original goals. It shows that RNAFlow has the potential to design new RNAs with certain features.
A glimpse into the future of RNA design
The possibilities that come with being able to make RNA are numerous and exciting, as shown below. Such prospects are:
- Development of novel drugs based on RNA: These RNAs can be manipulated so that they will interfere with defective proteins responsible for specific diseases. Therapeutic RNAs can be discovered faster using RNAFlow.
- Biosensors fabrication: Certain body-responsive RNAs can be engineered for molecule sensing purposes. This could lead to better medical diagnostic facilities in terms of sensitivity and efficiency with the help of RNAFlow.
- Understanding how RNAs work: By designing these molecules this way, scientists may learn more about cellular processes in a detailed manner.
Challenges and Looking Ahead
Despite the above advantages, there is still something that can be improved. Flexible RNAs are not an easy fit for the current version, as a result. Then again, the accuracy of protein-RNA interaction prediction methods restricts the design of RNAs that rely heavily on protein interactions to work.
These limits are recognized by authors who suggest solutions for them. If only protein-RNA folding and docking models could become more precise within RNAFlow’s capabilities, achieving those goals for which molecular recognition leads to rational design would be enhanced significantly. More work on RNAFlow would revolutionize the field of RNA design, opening doors to new therapies based on RNA and research tools.
Join The Conversation
The emergence of RNAFlow testifies to how instrumental artificial intelligence can be applied in the bioinformatics field.
What do you think about this novel way to create RNA? Will it transform the future of medicine and research? Leave your thoughts in the comments below!
Let’s talk about what lies ahead in RNA design and its endless possibilities!
Article Source: Reference Paper | RNAFlow code is available on GitHub.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.