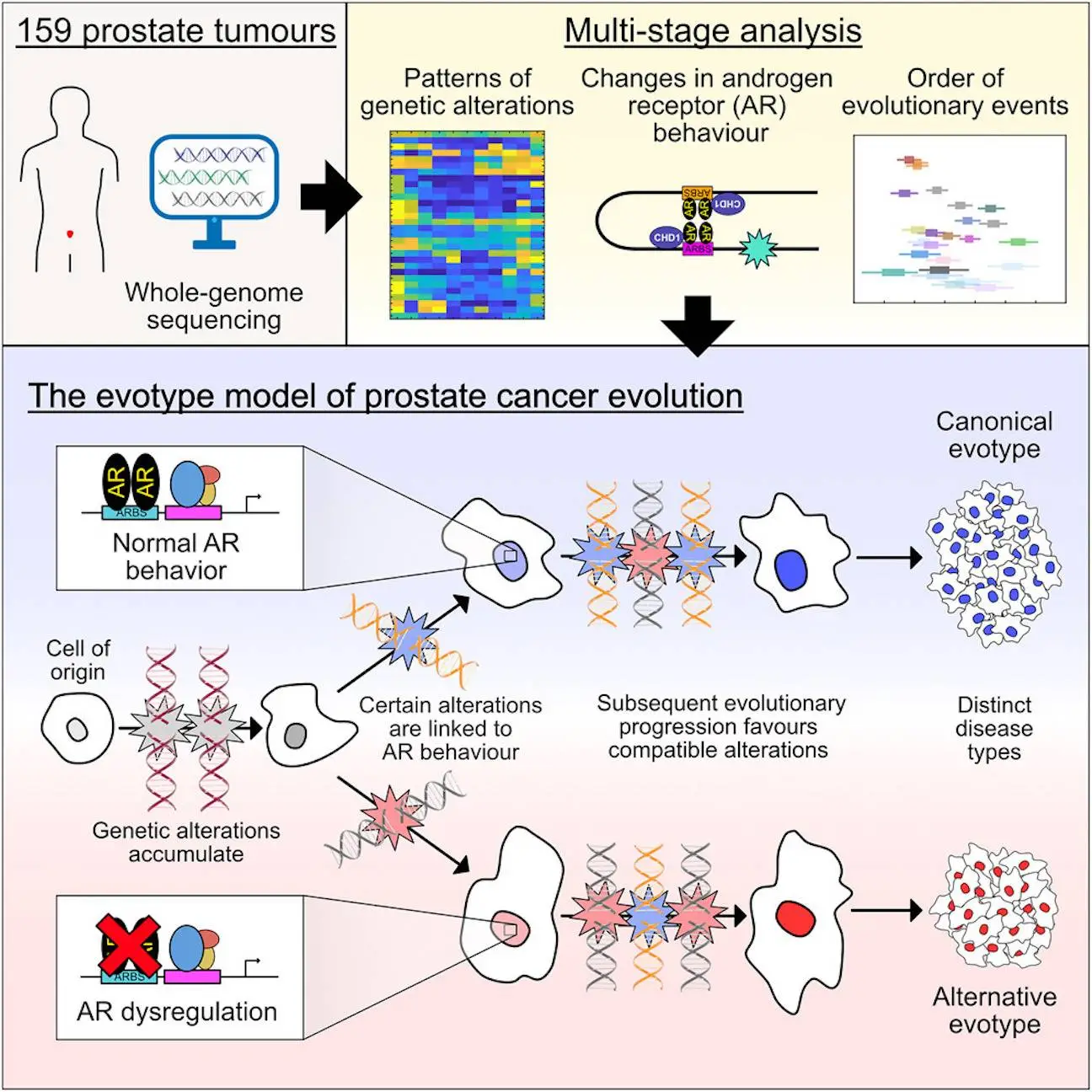
Prostate cancer remains a significant health threat for men globally. The development of cancer is an evolutionary process involving the sequential acquisition of genetic alterations that disrupt normal biological processes, enabling tumor cells to proliferate and ultimately invade and metastasize to other tissues rapidly. New research sheds light on how a prostate cancer diagnosis can affect one’s physical, emotional, and mental well-being, along with the environment that shapes prostate cancer evolution. The study was funded in part by Prostate Cancer Research and involved scientists from the University of Oxford, the University of Manchester, the University of East Anglia, and the Institute of Cancer Research in London. The researchers investigated the genomic evolution of prostate cancer through the application of three different classification methods, each designed to study various aspects of tumor evolution. Integrating the results revealed the existence of two distinct types of prostate cancer that arise from divergent evolutionary trajectories, designated as the Canonical and Alternative evolutionary disease types. Thus, they propose the evotype model for the development of prostate cancer, in which tumors of the Alternative-evotype deviate from those of the Canonical-evotype due to the random accumulation of genetic alterations associated with abnormalities in the binding of the androgen receptor. This blog dives into a recently published study that unveils a novel framework for understanding this disease.
Introduction
The dynamic process of tumor development involves the accumulation of genetic changes that cause abnormal phenotypes by interfering with normal cellular activities. While certain tumors may be divided into subtypes, typically employing strong genomic or transcriptome variations, the evolutionary mechanisms that give rise to this heterogeneity are complicated and not well understood. However, it has been shown that the order of events in some hematological malignancies can be related to prognosis and treatment susceptibility. Subtyping approaches for prostate cancer have been proposed based on gene expression patterns, specific molecular abnormalities, or combinations of changes. Nonetheless, extensive research undertaken by researchers has revealed substantial tumor heterogeneity, making it challenging to define subtypes in a simple or trustworthy way. The evolutionary factors that drive the emergence of prostate cancer subtypes remain largely unexplored. Simply put, existing methods for subtyping struggle to consistently categorize tumors.
The role of “evolutionary timing” in prostate cancer subtypes has been examined in a recent study. These investigations have shown that the order in which genetic alterations occur may, in certain cases, influence the course of the disease and its response to treatment. Every tumor has a unique evolutionary fingerprint. To investigate how evolutionary behavior manifests in the variation observed in prostate cancer genomes, researchers performed three separate analyses, each of which probes different aspects of tumor evolution. They did an unsupervised categorization of the malignancies in each analysis and subsequently identified tumor categories that were consistent across all investigations. Using this strategy, we may identify tumors that display consistent evolutionary properties and use this information to identify likely mechanisms driving prostate cancer evolution.
A Tale of Two Paths
According to the research, different forms of prostate cancer arise from random evolutionary processes that follow different paths and may be influenced by single genetic mutations that shift the balance in favor of convergence in either direction. Unlike the evolution of species, which involves ongoing adaptation to a perpetually changing environment, tumor evolution has a definable endpoint—a disease state that leads to the death of the host. As a result, more “evolved” cancers are closer to this target, which has obvious implications for risk assessment. Thus, it was proposed that the researcher’s evolutionary model implied two factors associated with risk: the evotype itself and the degree of progression relative to that evotype. This new perspective offers exciting possibilities for improving risk stratification, prognosis, and treatment strategies.
A New Framework for Understanding Prostate Cancer
Using integrated analysis, tumors were divided into two evotypes:
- Canonical-evotype (n=125):
DNA breaks are more affected by AR binding, indicating a traditional evolutionary pathway. Genetic alterations to the Androgen Receptor (AR), a protein required for prostate cell growth, appear to be at the root of this. Prostate cancer is frequently related to alterations in the function of the Androgen Receptor, which acts as a control switch for prostate cells.
- Alternative-evotype (n=34):
The second group appears to deviate from the Canonical route. Surprisingly, anomalies in AR DNA binding seem to have a secondary role in characterizing these tumors, which seem to be characterized by a unique set of genetic alterations. AR dysregulation can transpire without reference to AR binding sites, implying an alternative evolutionary pathway.
Although a unique defining mutation for the Alternative-evotype could not be identified, researchers investigated other possible sources of AR dysregulation. They identified correlations between a lower occurrence of DSBs around relevant ARBS pairs and the deletion of a specific gene (CHD1) involved in AR binding. This implies that the deletion of CHD1, by diminishing DSBs linked to the DNA loop mechanism, might play a role in the Alternative-evotype.
Epidemiological implications might result from evotype classification. For instance, non-White racial groups display an increased incidence of many Alternative-evotype aberrations and may therefore have a higher predisposition for this disease type. Conversely, cancers arising in younger patients have enrichment for ARBS-proximal breakpoints and are reported to develop via a similar evolutionary progression to the Canonical-evotype. It could also be possible to tailor treatment plans to the specific evotype. It has been shown that cancers with anomalies identified more frequently in the Alternative-evotype are more susceptible to ionizing radiation, respond better to PARP inhibitors, and respond better to androgen ablation.
The Road Ahead
The work published today by this global consortium of researchers has the potential to make a real difference to people affected by prostate cancer. The more we understand about cancer the better chance we have of developing treatments to beat it. We are proud to have helped fund this cutting-edge work, which has laid the foundations for personalized treatments for people with prostate cancer, allowing more people to beat their disease.
— Dr Rupal Mistry, CRUK’s senior Science Engagement Manager
The evolutionary model of prostate cancer disease types provides a conceptual framework that incorporates the findings of multiple previous studies and has significant implications for our understanding of the illness’s progress, probability, and therapy. As evolution through the sequential acquisition of synergistic genetic alterations is a process common to many tumors, the principles, analytical approach, and conceptual framework outlined here are widely applicable, and researchers anticipate them leading to insights into disease behavior in other cancer types.
The evotype model represents a paradigm shift in understanding prostate cancer evolution. It opens doors for:
- Developing more targeted therapies
- Optimizing existing therapies
- Identifying novel therapeutic targets
While the study presents compelling evidence for the existence of at least two distinct evolutionary pathways in prostate cancer, which forms the basis for classifying these cancers into evotypes. However, the precise criteria that differentiate Canonical-evotype tumors from those of the Alternative-evotype remain to be rigorously defined. The researcher’s statistical classification may, therefore, have incorrectly assigned some tumors to an evolutionary path that does not reflect their true nature. Additionally, the study population may not be fully representative of the global prostate cancer landscape. Despite these, the evotype model provides a solid framework for future research. The analytical technique might be utilized to treat other types of cancer, resulting in a better understanding of the illness’s behavior and, ultimately, improved patient outcomes. The struggle against prostate cancer is far from ending, but this study is a significant step closer to treating this challenging ailment.
Article source: Reference Paper | Reference Article | Processed data and code used in the study are available on GitHub
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.