Centuries of scientific invention have been driven by the search for new drugs to fight against diseases. Since medicinal plants became valuable sources of medicine from synthetic chemistry, we have consistently improved our capacity to design molecules with therapeutic potential. However, the traditional drug discovery process is time-consuming, costly, and fraught with challenges. It is here that ETH Zurich Chemists develop DRAGONFLY (Drug-target interActome-based GeneratiON oF noveL biologicallY active molecules), a novel de novo drug design method, promising to make the creation of new therapeutics easier.
Hurdles in Traditional Approaches to the Quest for New Drugs
The conventional drug discovery pipeline is a slow process involving target identification, hit discovery, lead optimization, and pre-clinical and clinical development. It has produced numerous drugs that have saved millions of lives, but there are limitations.
- Sequential Nature: The conventional drug discovery pipeline is a slow process involving target identification, hit discovery, lead optimization, and pre-clinical and clinical development. It has produced numerous drugs that have saved millions of lives, but there are limitations.
- High Attrition Rates: Inefficacy, side effects, or unfavorable pharmacokinetic properties result in most candidate drugs being eliminated from the pipeline. With such high attrition rates, huge financial investments are made without any guarantee of success.
- Limited Exploration: Screening compound libraries consisting of known compounds or natural products is the mainstay of these methods, thereby missing out on an entirely new chemical space with potential drug candidates.
De Novo Drug Design – Paradigm Shift
De novo drug design revolutionizes drug discovery. It entails using computer programs to create molecules from scratch for specific medical purposes. Its novelty lies in the following:
- Curing Serendipity: De novo design allows researchers to skip the accidental discovery of lead compounds and start designing molecules with specific properties.
- Expanding Chemical Space: On the frontiers of chemical space, de novo design leads us to unknown areas where we may find new classes of medicines with higher efficacy and fewer side effects.
- Speeding up drug discovery: De novo design may considerably speed up this process by making first stages more efficient, thus cutting down time and resources needed for bringing out new treatments for patients.
DRAGONFLY: A Fantastic Machine Learning Tool for De Novo Drug Design
The DRAGONFLY technique is a major advancement in de novo drug design. It employs deep learning, which is part of artificial intelligence (AI), to create new drugs. What makes DRAGONFLY distinct?
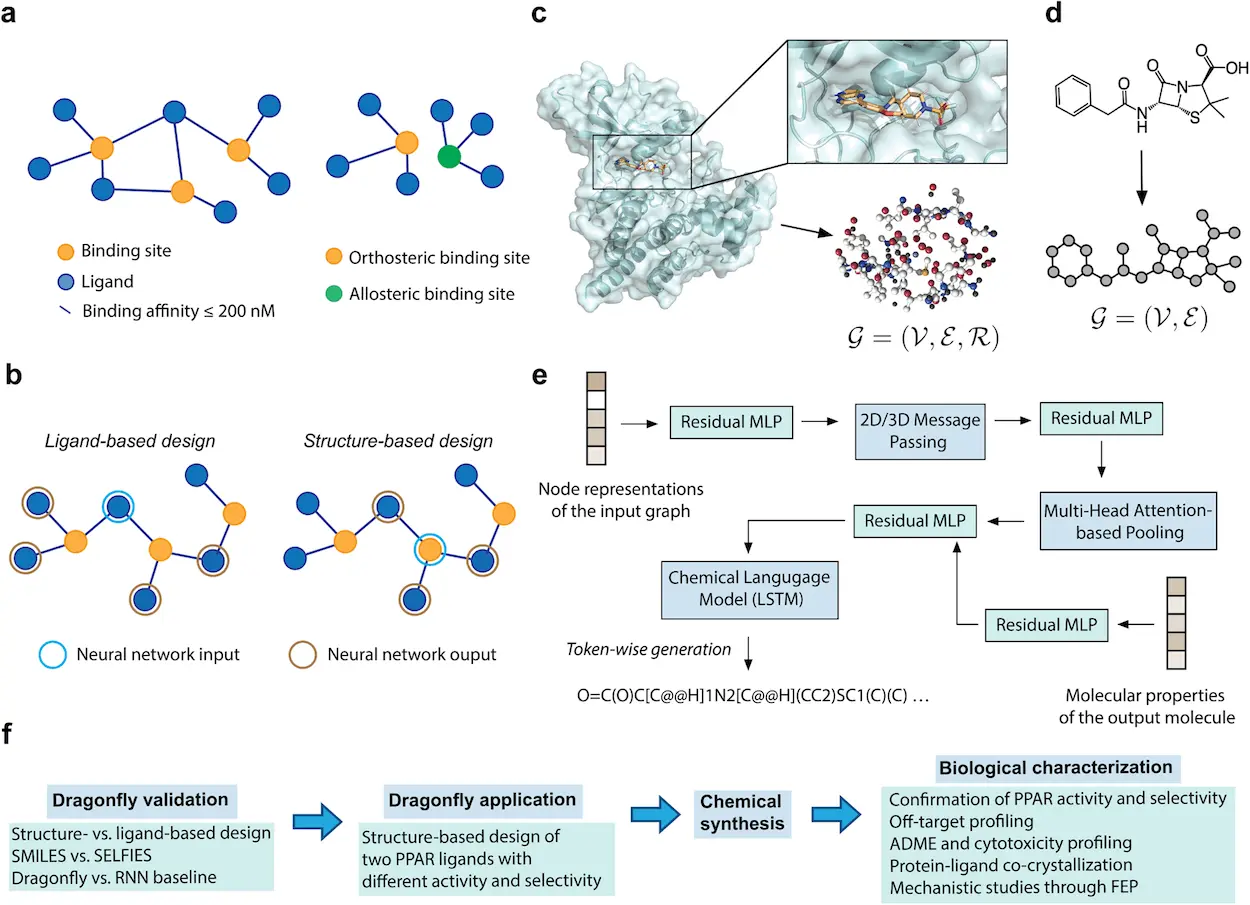
- Interactome Powerhouse: Unlike conventional methods that focus on individual molecules, the concept of interactome was adopted by DRAGONFLY. This network helps DRAGONFLY analyze and understand drug-target interactions better, thus building biologically relevant molecules that fit seamlessly into these pathways.
- Dual-Modality Champion: DRAGONFLY uses two different modalities to generate new drug candidates: ligand-based and structure-based design.
- Ligand-Based Design: In this approach, the known ligand is provided to the DRAGONFLY molecule, which binds specifically to a protein target, where it looks through the interactome, identifies novel compounds with similar properties, and is predicted to bind to the same protein target.
- Structural Design: The input for DRAGONFLY here is the three-dimensional structure of a protein’s binding site. Then, it uses deep learning skills to create molecules that correctly fit into the pockets and could act as antagonists or agonists, depending on the desired therapeutic effect.
- Transfer Learning: One major advantage of using DRAGONFLY is that transfer learning- an approach widely used in deep learning where models are trained on large collections of existing drugs- will not be necessary. This reduces the design task and allows DRAGONFLY to extend into areas that have never been reached within chemical space without being confined by known drug limitations.
Putting DRAGONFLY to the Test
The ultimate test of any scientific innovation lies in its ability to show material outcomes. They applied their technique through stringent validation processes to a real-life problem – designing ligands that target human peroxisome proliferator-activated receptor gamma (PPARγ). Type 2 diabetes and metabolic disorders are among the conditions associated with this protein.
The scientists in this study used the structure-based design approach of DRAGONFLY and fed the three-dimensional structure of the PPARγ binding site into the model. The model then successfully produced a library featuring all-new drug candidates. After an intensive laboratory evaluation, some potential promising leads were identified and studied further. These molecules showed potent PPARγ partial agonist activity, which expressed desired selectivity profiles and favorable ADME characteristics. Remarkably, crystal structure determination showed that the top-ranked compound was bound to PPARγ in exactly the orientation predicted by DRAGONFLY. This incredible convergence between virtual and physical worlds constituted a strong proof of concept for the model.
The Road Ahead: Challenges and Opportunities
Many researchers are now exploring its potential in making drugs against a broader range of therapeutic targets. However, there are exciting possibilities:
- Targeting Previously Untreatable Diseases: There are many diseases without effective cures because of the limitations of traditional drug discovery methods. Hence, DRADRAGONFLY’Sility to explore unexplored chemical spaces could help discover new therapeutic targets for previously untreatable conditions.
- Personalized Medicine: The ultimate goal of medicine is to be able to create custom treatments based on the exact genetic composition of an individual. It is, therefore, possible that through specific target-oriented drug development by DRAGONFLY, personalized medical approaches will have a better outcome in terms of their efficiency and focus.
- Repurposing Existing Drugs: Therefore, DRAGONFLY might be used to find novel applications for already existing medicines. Thus, by considering its interactome alongside its structure, the model could predict other potential targets with which it can interact, thereby suggesting new therapeutic uses for currently known drugs.
The long and winding road to the widespread adoption of DRAGONFLY is still marked with some challenges:
- Data Integration: DRAGONFLY hinges on the availability of a great deal of information, including protein structures, drug interactions, and disease biology. Integrating and managing this data effectively is key to furthering the model.
- Experimental Validation: The predictions must be tested extensively in laboratories to ascertain their accuracy. While the PPARγ case study provides some optimism, more experiments must be done to cement its trustworthiness.
- Integration into Drug Discovery Pipelines: AI researchers must work closely with medicinal chemists and pharmaceutical companies to fit smoothly into prevailing drug discovery workflows.
Conclusion: A New Era of Drug Discovery
The deep learning and interactome analysis-based DRAGONFLY represents a shift from how new drugs are designed. This innovation can help us enter a new era of drug design that is fast and efficient and covers virgin grounds within the chemical space. So, as DRAGONFLY undergoes further enhancements, it will lead to faster, much-targeted development of life-saving drugs that could revolutionize healthcare for generations to come.
Article Source: Reference Paper | Code availability: GitHub | Zenodo
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.