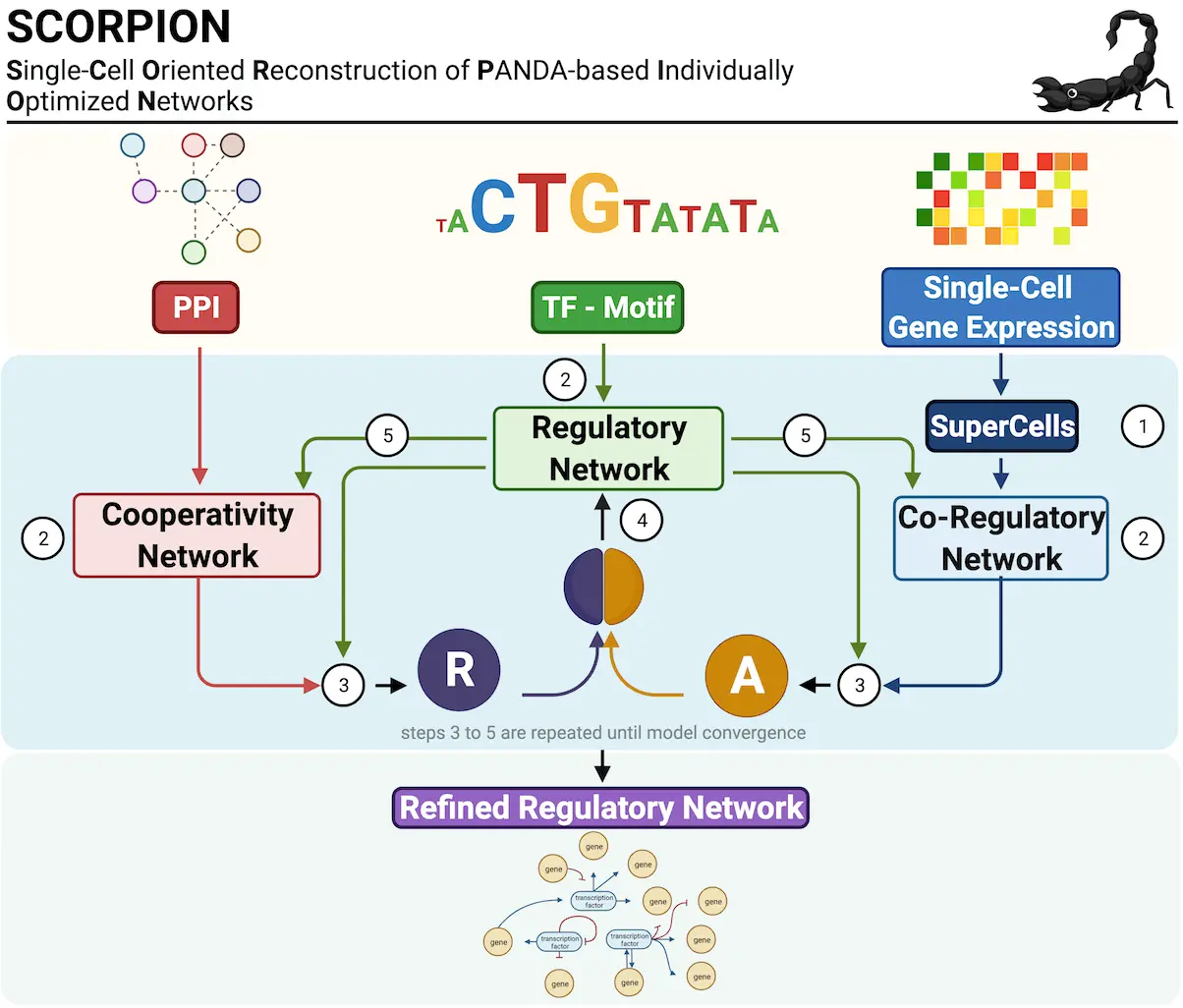
Gaining insight into the intricate workings of genes within cells is essential to comprehending biological processes and illnesses. Bulk RNA sequencing, which hides diversity within cell populations and analyzes average gene expression across numerous cells, has historically been the focus of study. On the other hand, single-cell RNA sequencing, or scrRNA-seq, has become a powerful technique for examining gene expression in individual cells. Improved understanding of cellular diversity and gene regulation results from this fine-grained resolution. A new paper from the University of Texas at Austin researchers introduces SCORPION, a one-of-a-kind R program tool that generates GRNs (gene regulatory networks) from scRNA-seq data with remarkable precision and efficiency. SCORPION differs from prior techniques in that it incorporates more than just gene expression data sources. SCORPION outperformed 12 existing gene regulatory network reconstruction techniques. SCORPION will help explore crucial aspects of precision medicine, health, and biomedical research.
The Challenge of Single-cell Network Analysis
In eukaryotes, transcription factors—proteins required for cell identity and state management—carefully control gene expression. They achieve this by regulating or boosting the expression of specific target genes. The number of transcription factors, their ability to bind to chromatin (a DNA-protein complex), and the many post-translational alterations they undergo all influence this regulation. Gene regulatory networks for every kind of cell or condition within a single sample may be inferred from the gene expression variability seen in RNA-sequencing (RNA-seq) data from single cells/nuclei.
Sparseness is a common feature of scRNA-seq data, meaning that little is known about each gene in each cell. It is challenging to identify minute correlations between genes that suggest regulatory connections because of this sparsity. Moreover, single-cell data may contain cells at different phases, which might complicate the research. Current methods sometimes suffer from sparsity and don’t take into consideration the underlying variability inside individual cells.
Introducing SCORPION
To address these challenges in differential gene regulatory network analyses on single-cell data, scientists present SCORPION (Single-Cell Oriented Reconstruction of PANDA Individually Optimized gene regulatory Networks), a tool that uses coarse-graining of single-cell/nuclei RNA-seq data to reduce sparsity and improve the ability to detect correlation structures in these data. This program tries to overcome these issues in differential gene regulatory network analysis using single-cell data. The regulatory network reconstruction approach (PANDA) is then used to reconstruct gene regulatory networks based on the coarse-grained data generated. PANDA uses a message-passing approach that combines data from several sources, including sequence motif analysis, gene expression, and protein-protein interactions, to predict regulatory links. Owing to the coarse-graining and the use of the same baseline priors for each aggregated Super/MetaCell, SCORPION can reconstruct comparable, fully connected, weighted, and directed transcriptome-wide gene regulatory networks suitable for statistical analyses that leverage multiple samples per experimental group—as ‘population-level studies.’
Researchers tested the performance of SCORPION’s coarse-grained input data for network modeling using synthetic data via BEELINE, a tool for systematically evaluating cutting-edge algorithms for inferring gene regulatory networks from single-cell transcriptional data. They found that networks modeled on data desparsified with SCORPION outperformed 12 other gene regulatory network reconstruction techniques across seven metrics. In addition, using supervised experiments, they showed that SCORPION can precisely identify biological differences in regulatory networks between wild-type cells and cells carrying transcription factor perturbations. Furthermore, they used SCORPION to analyze a single-cell RNA-seq atlas constructed from publically accessible data, which comprises 200,436 cells from 47 individuals and covers three unique areas of colorectal cancers and healthy surrounding tissue. This highlights SCORPION’s scalability to population-level analyses.
The SCORPION investigation revealed variations between the intra- and intertumoral zones, which aligns with our understanding of the mechanisms by which the chromosomal instability pathway (CIN), responsible for most colon cancer cases, drives the advancement of the illness. Findings were confirmed in an independent cohort of patient-derived xenografts from left- and right-sided tumors and provided insight into the regulators associated with the phenotypes and the differences in their survival rate.
Exploring SCORPION’s Applications
SCORPION’s versatility was proved by researchers who used it to examine actual scRNA-seq data from many studies:
Studying the roles of specific transcription factors:
It examined real single-cell RNA-seq data from studies involving the transcription factors DUX4 and Hnf4αγ.
Constructing a comprehensive scRNA-seq atlas of colorectal cancer:
Furthermore, a huge single-cell RNA-seq atlas of colorectal cancer was exposed to SCORPION, which enabled the following:
- The development of gene regulatory networks specific to cell types.
- Modeling tumor development using gene regulatory networks.
- Finding differences in cancer gene regulatory networks on the left and right sides.
Strengths and Benefits of SCORPION
- Provides data on transcription factor binding patterns, protein-protein interactions, and gene expression.
- During network development, motif footprints are used to build a more complete picture.
- Uses exact association metrics to evaluate the activity of undiscovered transcription factors.
- Sparse matrices decreased primary components, and CRAN availability improved computation performance.
- Allows for the analysis of single-cell data using typical statistical methods.
- Improves knowledge of molecular processes and enables precision medicine research.
The Road Ahead
SCORPION outperforms other gene regulatory network design tools in terms of computational efficiency. It employs sparse matrices by default, which saves memory and accelerates matrix multiplication. To boost computation performance, it additionally employs shorter primary components during the desparsification stage. Furthermore, SCORPION is widely available across several platforms thanks to the CRAN repository, making installation and use across a variety of operating systems easier.
SCORPION enables the use of statistical methods that account for population variation and are often used in other areas of genomic data analysis by generating precise and extremely comparable gene regulation networks for each sample. These methods include differential analysis, dimensionality reduction, and grouping based on sample similarity. Now that gene regulatory network perturbations have been shown to be effective at reproducing experimental results, and it is anticipated that SCORPION will be used to investigate a wide range of important questions in precision medicine, health, and biomedical research, in addition to characterizing the molecular mechanisms driving phenotypes.
Article source: Reference Paper | SCORPION code availability: Package | GitHub
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.