Next-generation sequencing has transformed gene expression profiling into a routine experiment, but interpreting those matrices remains a challenge for many labs. Researchers from Kyungpook National University, Republic of Korea, address this gap by developing an “agentic” AI system that can plan, execute, and interpret RNA-sequencing downstream analysis for researchers with limited biological backgrounds. As a bioinformatician, this feels less like yet another web tool and more like a serious attempt to give newcomers a competent co‑pilot for exploratory data analysis.
Why Downstream Analysis Still Hurts
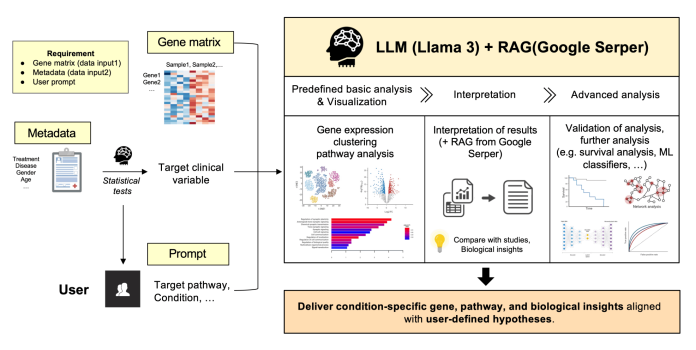
Building a count matrix is not the most time-consuming task; what’s slowing projects down is actually everything that follows. Steps like differential expression, clustering, and pathway analysis: each step requires an immense amount of statistical knowledge and coding, as well as knowledge of the specific domain of the task, and most wet-lab researchers simply do not have that. Existing agents in the field, like CellAgent and BioAgents, automate some of the workflows in the domain. However, these do not hold the hand of the user when it comes to guiding them on the interpretation and development of the hypothesis. The authors position their systems to fill in this gap, not only generating these graphs and plots, but narrating to them the biological mechanisms in the plots, guiding from there as to what the user should focus on next.
An Agentic Workflow, Not Just a Script
Upon receiving the gene expression values and clinical metadata in a .csv format and starting from an example cancer-enriched dataset(200 genes × 100 samples), the user gets to interact with a system that is deployed as a modular Python pipeline with a Streamlit frontend. The system normalizes the expression values by applying a log2 transformation, and clinical variables are screened via Pearson correlation or t‑tests to generate a concise significance table that flags which features are worth focusing on. The agentic part comes from the LLM(Llama 3 70B), which plans and executes the analysis sequentially, reflects on the intermediate outputs, and ensures alignment of the entire flow to what the user is comparing and what the question is.
From DEGs To Pathways And Plots
After choosing a main clinical characteristic, the system employs t-test differential expression, computes log fold changes, and determines which genes are significantly different. It then analyzes the different dimensions in the data using k-means clustering on the transposed expression matrix, and samples are projected in PCA space and colored by clinical characteristics (e.g., disease status). On top of this, it connects DEGs to biology using gProfiler and GSEApy to perform functional enrichment, summarizing the most important pathways in clear bar plots. These are then presented with a volcano plot and a PCA plot, which essentially automate the entire downstream workflow process into a guided, clickable interface.
Literature‑Aware Interpretation For Non‑Experts
What differentiates this model the most is in the interpretation step. Instead of leaving the users with a volcano plot, the model analyzes the DEG clusters and enriched pathways, then queries PubMed using an RAG pipeline for contextualizing. To ground the model’s explanation in literature, abstracts are embedded and stored in a FAISS vector database, which is selectively queried for literature-backed explanations. If the dataset contains upregulated oncogenes like BRAF and is from diseased samples, the system is capable of linking to immune dysregulation or poor prognosis, as the model integrates log fold changes and p-values to known disease mechanisms in a way a non‑bioinformatician can follow.
Recommending And Running Advanced Analyses
Instead of just doing descriptive analysis, the agent chooses three more sophisticated techniques for the particular dataset, like survival analysis with Cox proportional hazards techniques, or machine-learning classifiers. All of them come with explanations and ready-made Python codes, so the user chooses the method with Streamlit, runs it in the app, and gets a written update and files they can pull. In the example provided by the authors, the system detects 40 significant DEGs, discovers clusters connected with a particular disease state, and uses transcriptional shifts to DEGs to validate survival hypothesis modeling, including ratios of attitude and shifts for those transcriptional changes.
Democratizing NGS Analytics
From a bioinformatician’s perspective, this work is less about replacing analysts and more about lifting the floor for everyone else. The system, mixing Biopython, gProfiler, GSEApy, advanced mathematics, and a large language model to be integrated by an agentic system, does not ‘dumb down’ ease of use by the user for downstream analysis performed on NGS data, particularly RNA-seq. The authors mention future improvements, like drug response prediction or multi-omics integration. Nevertheless, even in this state, the AI agentic system lowers the threshold for hypothesis-driven interpretation of NGS data, freeing human experts to focus on the toughest questions rather than the first pass through a count matrix.
Article Source: Reference Paper
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.