Researchers from the Indian Institute of Technology, Madras, present iCOMIC, a tool for quickly analyzing genomic data. With iCOMIC, users can analyze whole genome and exome sequencing data as well as RNA-Seq data using several pre-configured workflows. Developed as an open-source, standalone tool characterized by a Python-based GUI and automated bioinformatics pipelines for DNA-Seq and RNA-Seq data analysis, iCOMIC (integrating the COntext of Mutations In Cancer) makes life easier for clinical researchers and biologists.
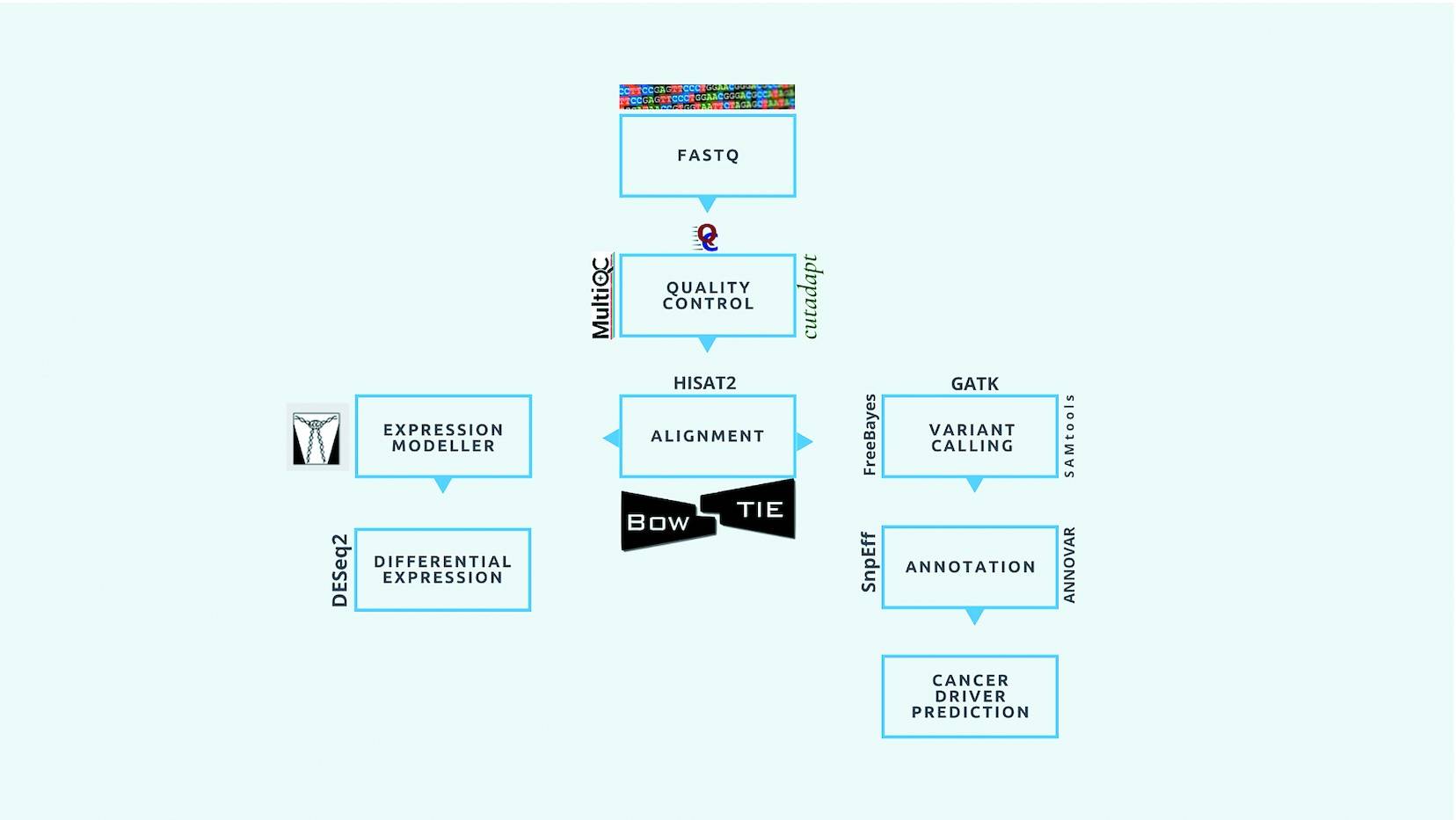
Although modern sequencing technologies generate tremendous amounts of omics data, analyzing them requires substantial bioinformatics expertise. In order to address this concern, a pipeline for the analysis of (cancer) genomic data that uses raw sequencing data (FASTQ format) as input and provides insight into the data based on its statistics was developed. iCOMIC toolkit pipeline, with many independent workflows, is integrated with the Snakemake workflow management system.
A user-friendly GUI makes it easy to analyze whole-genome and transcriptome data and eliminates the need for complex command-line arguments and minimal execution steps. From somatic mutation data, the researchers developed algorithms that predict pathogenicity among cancer-causing mutations and distinguish between tumor suppressor genes and oncogenes. Using the BWA MEM-GATK HC DNA-Seq pipeline was a benchmarked tool against the Genome In A Bottle benchmark dataset (NA12878) and achieved the highest F1 score of 0.971 and 0.988, respectively, for indels and SNPs. The HISAT2-StringTie-ballgown pipeline and the STAR-StringTie-ballgown pipeline achieved a correlation coefficient of 0.85 using the human monocyte dataset (SRP082682). This tool significantly improves complex data analysis pipelines by facilitating the analysis of large omics datasets.
The rise of Next-Generation Sequencing (NGS) technologies has greatly advanced genomic research over the past couple of decades. Researchers have discovered new DNA and RNA variants and differentially expressed genes as a result of these rapid advances in sequence-based analysis. RNA Sequencing (RNA-Seq) enables quantification of gene expression, while Whole Genome/Exome Sequencing (DNA-Seq) identifies nucleotide variants. Further, Whole Genome Sequencing is a powerful tool for analyzing mutations in cancer and is a cornerstone of personalized medicine. As a result of RNA-Seq, various biological phenomena can be further interpreted.
The massive amounts of data produced by NGS technologies have been analyzed using various bioinformatics tools. The need for an automated pipeline is exasperated by the difficulty of data analysis for biologists. The development of new tools for genomic data analysis occurs periodically, but there is no comprehensive toolkit. The use of different combinations of tools has been the subject of extensive comparative studies. A user-friendly toolkit that includes a surplus of bioinformatics tools that aids non-programmers is lacking, however, some software suites combine a few tools. The availability of open-source bioinformatics pipelines to analyze large (cancer) genomic datasets is also lacking.
What is iCOMIC?
In the paper published in NAR Genomics and Bioinformatics, the researchers introduced iCOMIC, a web-based platform that enables users to perform complex analyses of cancer genomic data using intuitive interfaces. Nonprogrammers may find it challenging to analyze the large amounts of data generated by Next Generation Sequencing techniques due to the computational skills required. Users with minimal programming skills will find iCOMIC to be a robust platform. With iCOMIC, users can analyze whole genome and exome sequencing data as well as RNA-Seq data using several pre-configured workflows.
A unique feature of iCOMIC is that it uses proprietary algorithms to predict cancer genes, tumor suppressor genes, and oncogenes, as well as cancer driver and passenger mutations. Software packages for Linux, Windows, and Mac can be downloaded as part of the iCOMIC toolkit. With iCOMIC, one can analyze data using an interactive GUI and install the software without any hassle. As a result of the above features, iCOMIC makes large genomic datasets easily accessible and open-source. Based on DNA-Seq benchmarking performed in iCOMIC, F1 scores for indels and SNPs were 0.971 and 0.988, respectively. The fold change correlation coefficient for RNA Seq was 0.85 when compared to a microarray dataset.
Users with minimal programming experience can use iCOMIC to analyze genomic data using a point-and-click application. It provides a user-defined combination of tools and a set of easily adjustable parameters for the analysis of genomic data, with a versatile, fully automated pipeline. A wide range of bioinformatics tools is integrated into iCOMIC to allow users to customize pipelines in less than five simple steps.
Features of iCOMIC
iCOMIC provides several features including:
- A suite of tools for performing basic analysis tasks (e.g., gene expression profiling)
- An interactive visualization tool for exploring data sets
- A set of prebuilt pipelines for performing advanced analyses (e.g., differential expression analysis, pathway enrichment analysis)
- A database system for storing and sharing results
- A web server for hosting the application and providing access to the data
iCOMIC vs. Galaxy
In Galaxy, when analyzing multiple samples, it is necessary to rename the files before passing them to the next tool in the pipeline, which can be tedious when there are a large number of samples. iCOMIC, on the other hand, automates the entire process. Galaxy is, without a doubt, a popular pipeline for analyzing genomic data, and it has many advantages. However, when it comes to simple data analysis, iCOMIC excels, and biologists and clinical researchers find it more appealing.
DNA-Seq and RNA-Seq data can be analyzed with iCOMIC as a standalone toolkit. Both germline and somatic variants can be detected using iCOMIC’s DNA-Seq component. The output of one tool is automatically transferred to the next in iCOMIC, unlike conventional analysis pipelines like Galaxy. Users with minimal programming experience are welcome to utilize iCOMIC’s interactive and user-friendly GUI. In addition, iCOMIC allows expert bioinformaticians to incorporate additional tools and advanced parameters into their analyses, saving time on pipeline development.
Python wrapper scripts connect Snakemake workflows to the GUI. A variety of tools can be selected from the predesigned combinations according to the user’s needs. In designing these individual workflows, the best connectivity between the tools has been considered. Modules can be replaced or pipelines altered with the help of iCOMIC. Additionally, tools and dependencies are easily installed using the conda environment.
Final Thoughts
iCOMIC is a robust platform for users with minimal programming skills. iCOMIC can be used to perform a variety of data processing, analysis, and transformation tasks of cancer omics data. iCOMIC enables the user to choose from several pre-configured workflows for analyzing Whole Genome/Exome Sequencing and RNA-Seq data. It also integrates novel algorithms developed by researchers to predict cancer driver and passenger mutations, as well as tumor suppressor genes and oncogenes. The iCOMIC toolkit can be downloaded as a package and run on Linux, Windows, or Mac operating systems.
Article Source: Reference Paper | iCOMIC source code: GitHub | iCOMIC user manual: Manual
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Srishti Sharma is a consulting Scientific Content Writing Intern at CBIRT. She's currently pursuing M. Tech in Biotechnology from Jaypee Institute of Information Technology. Aspiring researcher, passionate and curious about exploring new scientific methods and scientific writing.