An integrated atlas of the human 4D genome just dropped, and it is breathtaking in its ambition. In a massive consortium effort, researchers from various institutions, including UMass Chan Medical School, Northwestern University, and other collaborators, have assembled one of the most comprehensive views to date of how the human genome folds and functions in space and time inside the nucleus.
From linear genome to living 4D structure
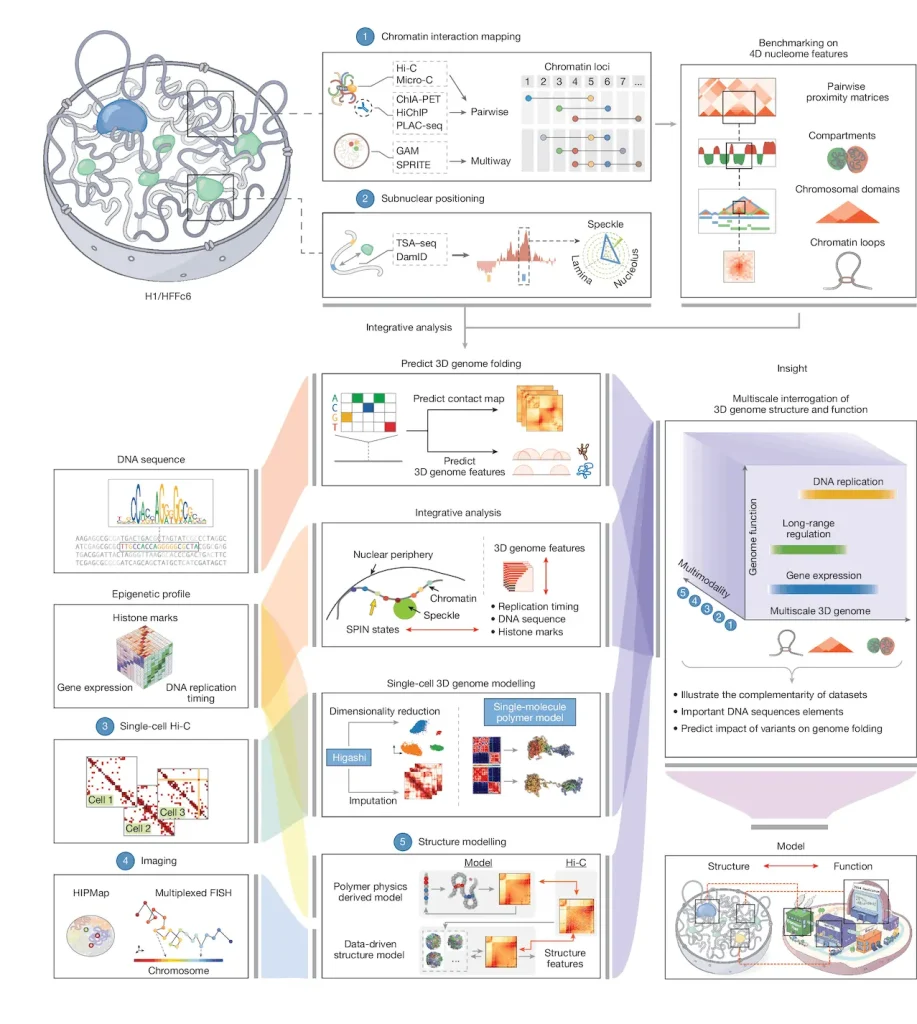
Image Source: https://doi.org/10.1038/s41586-025-09890-3
As bioinformaticians, genomes frequently consist of nothing more than a FASTA file, a few BED tracks, and some contact matrices. This research illustrates that genomes have materiality: a 2-meter-long polymer, folded into a ~10-micrometer nucleus, the 3D structure of which is intricately intertwined with the processes of transcription, replication, and the organization of nuclear bodies.
The consortium studied two “workhorse” human cell types: H1 embryonic stem cells and immortalized fibroblasts HFFc6. For each, they created and incorporated a substantial variety of assays: Hi-C, Micro-C, SPRITE, GAM, ChIA-PET, PLAC-seq, DamID, TSA-seq, Repli-seq, RNA-seq, iMARGI, and a few more. They then used deep machine learning to derive and Domestically Engineered genome-wide 3D (D3) Structures and “SPIN states,” which are spatial annotations describing the positional relationship of each locus to the lamina, speckles, nucleoli, and other nuclear compartments.
SPIN States Reveal Hidden Layers
The SPIN framework is an example of the application of the A/B compartment model, offering a unique perspective on genomic compartments by assigning each 25-kb genomic bin a unique spatial classification: Speckle-proximal, Interior Active, Near-Lamina, or Lamina-bound. Also incorporating the TSA-seq distances to nuclear structures, DamID lamina profiles, Hi-C contacts, and replication timing, these states demonstrate the ability to predict behavior with striking accuracy. Active speckle and interior zones replicate early and are transcriptionally active, while lamina regions replicate late in S-phase and are transcriptionally inactive. SPIN provides further granular detail to classic A/B compartments by illustrating active zones or transcriptionally active interiors against a backdrop of transcriptional silence.
This lends itself to the creation of an empirical annotation track, similar to ChromHMM but enhanced with 3D contextual information, which can then be overlaid with GWAS signals, enhancers, or regulatory maps for a more nuanced understanding of spatial regulation.
Loops and Enhancers Power Gene Expression
Patterns can be recognized in a union catalog that has more than 140,000 loops for each cell type. Genes associated with more distal enhancers tend to have higher expression levels, and the number of enhancers predicts expression in 116 different tissues, with sparse connections for specialist genes and plentiful connections for workhorse genes. Housekeeping genes surprise with over 90% of them connecting to at least one enhancer in both cell types, often many, with 80% of the loops switching dynamically between stem cells and fibroblasts to maintain a constant level of expression.
Even some genes that are trapped at the nuclear lamina can use enhancer loops in active pockets, which are regions that loop out of the repression. The scale of the structure is important; megabase compartments that coincide with the replication domains are the size of nested TADs that do not override each other, and the centers of replication origins congregate at the edges of ‘dot’ TADs, suggesting a relationship between the timing of DNA replication and gene extrusion.
AI Unlocks Variant Predictions
The study culminates in deep learning models trained on Micro-C data, simulating how motif mutations disrupt loops. Stem cell models sensitively react to SOX2 perturbations, fibroblast ones to FOS/JUND tweaks. Explainable AI pinpoints sequence drivers, opening doors to predict how disease variants might scramble 3D architecture beyond mere binding changes.
This 4D nucleome atlas integrates disparate datasets into a single, readily accessible resource via the 4D Nucleome portal. It will power the next stage of genomics research.
Article Source: Reference Paper | Reference Article | Code Availability: GitHub
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.