The digestive system is home to trillions of bacteria, which are very different in each person based on their gender, age, race, lifestyle, and health. For precision medicine to include microbial metabolism, computer models would have to be expandable and able to resolve strains and molecules. This study introduces us to AGORA2 – assembly of gut organisms via reconstruction and analysis, version 2. The strains in AGORA2 were carefully chosen using comparative genomics and literature searches. The strain-resolved drug degradation and biotransformation capabilities for pharmaceuticals were also considered.
The COBRA approach
Several commonly prescribed medications work better and are safer when the human microbiota is healthy. It has been shown that microbes in the human gut metabolize 176 of the 271 medicines that have been looked into. This activity differs from individual to individual. So, in precision medicine, treatments have been suggested that use nutrition, genetics, and the microbiome. Constraint-based reconstruction and analysis (COBRA) is a mechanistic approach to systems biology that uses a precise stoichiometric representation of metabolism. COBRA uses genome-scale reconstructions of the target species that were chosen by hand based on the available data.
With the help of condition-specific constraints, such as metagenomics and nutritional data, these reconstructions can be turned into predictive computational models and linked to strain-resolved, customizable microbiome models. So, the COBRA method is a good way to look into cometabolism in the human microbiome. CarveMe, MetaGEM, MIGRENE, and gapseq are some of the semiautomated reconstruction technologies that have been developed to help reconstruct the hundreds of known species that live in humans at the genome-scale. Even though these tools have a lot of advantages, they don’t help much with curation against manually improved genome annotations and experimental data from peer-reviewed literature.
Enhancement of AGORA: AGORA2
AGORA2 is an expansion of AGORA’s scope and coverage. It includes reconstructions of microorganisms for 7,302 strains, 1,738 species, and 25 taxa. More than 5,000 strains, 98 compounds, and 15 enzymes, some of which have been confirmed by experimental data, have been added to AGORA2 as a hand-designed molecule- and strain-resolved drug biotransformation and degradation processes. The AGORA2 reconstructions are in line with the whole-body, organ-resolved, generic, and sex-specific human metabolic reconstructions.
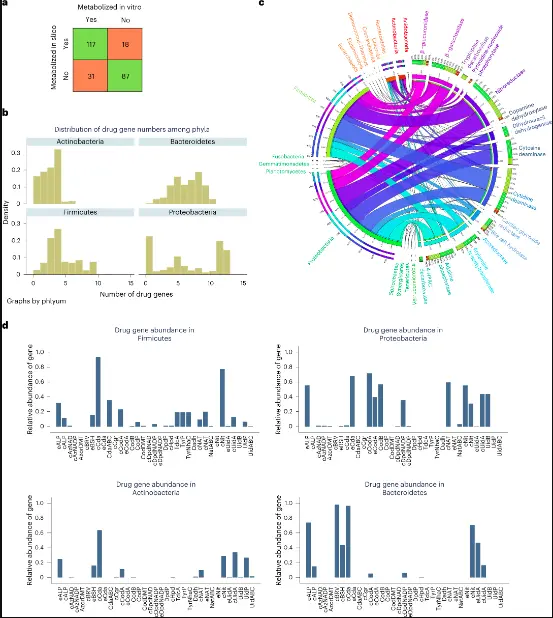
Image Source: https://doi.org/10.1038/s41587-022-01628-0
Data-driven reconstruction
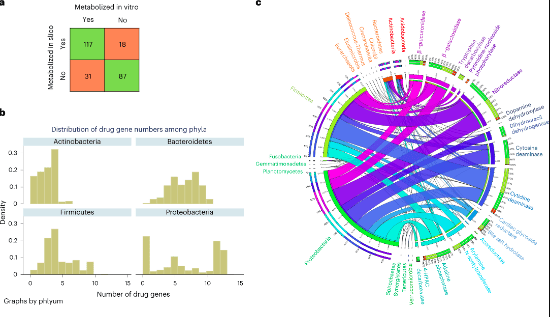
A data-driven reconstruction approach called DEMETER (Data-drivEn METabolic nEtwork Refinement) was enhanced in order to generate reconstructions of the 7,302 gut microbial strains in the AGORA2 database. The DEMETER procedure encompasses data collecting, data integration, draught reconstruction development, translation of reactions and metabolites into the Virtual Metabolic Human (VMH) namespace, and concurrent iterative refinement, gap-filling, and debugging. As the absence of accurate genome annotations is a source of uncertainty in the prediction ability of genome-scale reconstructions, the annotations of 446 gene functions across 35 metabolic subsystems for 74% of the genomes were manually validated and improved. To further ensure an accurate portrayal of species-specific metabolic capacity, a comprehensive literature search yielded data for 95% of the strains from 732 peer-reviewed papers and two microbial reference textbooks. Due to these curation efforts, metabolic models derived from the improved reconstructions showed a substantial improvement in their ability to forecast compared to models derived from KBase draft reconstructions. The contents of AGORA2 reconstructions were then categorized based on their taxonomic distribution. AGORA2 demonstrates the diversity of the gathered strains by grouping them by class and family in line with response coverage.
AGORA2 and three independent datasets
Further testing of the quality of AGORA2 and the DEMETER pipeline was conducted by comparing AGORA2’s prediction capabilities and model characteristics to those of existing resources for genome-scale reconstructions of microorganisms. To achieve this objective, 8,075 gapseq reconstructions, 1,333 MIGRENE reconstructions labeled as MAGMA, and 72 manually chosen genome-scale reconstructions were added to the BiGG database. For an objective assessment of the reconstruction’s quality, the fraction of flux-consistent reactions in each resource was calculated. Only manually curated BiGG and CarveMe reconstructions had a higher percentage of flux-consistent reactions than AGORA2. The most essential aspect of a genome-wide reconstruction is its accuracy in capturing known biochemical or physiological features of the target organism. The AGORA2 reconstructions impressively captured the known metabolite absorption and secretion capacities of the target species.
To directly compare the results of AGORA2 with KBase, CarveMe, gapseq, BiGG, and MAGMA, the uptake and secretion accuracy of each model were measured independently. Using a nonparametric sign rank test, the precision of models was evaluated in the overlap between AGORA2 and each resource. AGORA2 outperformed all other methods on all three datasets, except for BiGG on the BacDive dataset, for which the overlap between models was inadequate to achieve sufficient statistical power. Collectively, the AGORA2 reconstructions accurately represent the known properties of the diverse species, exceeding earlier semiautomatically generated reconstructions and rivaling manually picked reconstructions. As a result, AGORA2 scored very high for metabolite absorption and secretion data, which need curation based on experimental data, compared to enzyme activity data, which can be verified based on genomic annotations.
Drug metabolism in microbes and genomics
Microorganisms may have direct or indirect effects on the way a drug works and how toxic it is through degradation and biotransformation. Genome annotation methods only give a limited picture of how drugs are broken down, and no one has ever done a full comparative genomic study of enzymes that break down drugs. So, there are currently no genome-scale reconstruction resources that can take into account how microbes change drugs. To fix this problem, a thorough manual comparative genomic study was done. It looked at 25 drug genes, 15 enzymes that have been shown to directly or indirectly change drug metabolism, their subcellular locations, and 12 drug transporter genes. Based on genetic evidence, the reconstructions included an average of 188 drug-related reactions, 111 metabolites, and 1,440 drug-related reactions. This was proven with a precision of 0.81.
Personalized modeling of drug-metabolizing capacities
Human microorganisms do not occur in isolation, so the essential question of how the total drug-metabolizing ability may differ amongst gut microbiomes was discussed. A metagenomic dataset from a Japanese cohort of 365 patients with colorectal cancer (CRC) and 251 healthy controls was used to study the metabolic capacities of each gut microbiome and confirm the fluxes against metabolomic data. 97% of the species that were found could be mapped into AGORA2, while only 72% could be mapped into AGORA. For each person’s gut microbiota, a community model was made and analyzed. This led to a prediction of the person’s overall capability to break down drugs.
Drug-metabolizing capacities and clinical parameters
Next, the connection between the ability to metabolize drugs and CRC (Colorectal Cancer) was examined. Even though CRC metagenomes were found to have more than 29 species, neither the quality nor the amount of drug-metabolizing ability changed for any of the medicines, including those used to treat cancer.
The statistical relationship between age, gender, and body mass index (BMI), and the ability of the microbiome to break down drugs were also looked at. There was a clear link between age and five of the predicted secretion potentials of drug metabolites, but the effect sizes were small to medium. In conclusion, the study made it possible to look at clinical factors related to the ability of the microbiota in the gut to break down drugs.
AGORA2-based community modeling
In the last part of the validation process, whether AGORA2-based community modeling can predict the sign of statistical relationships between the presence of microbial species and the concentrations of fecal metabolites in the CRC sample was examined using methods that have already been published. The fact that AGORA2-based community models can predict the direction of species-metabolite correlations for a wide range of metabolites shows that the models are good at making predictions.
Conclusion
AGORA2 is a set of 7,302 genome-scale reconstructions for human-associated microbes that, as far as we know, have never been done before in terms of coverage, breadth, and curation effort. AGORA2 meets the quality standards set by the systems biology research community, describes the biochemical and physiological traits of the target organisms more accurately than other reconstruction resources, and includes drug-metabolizing abilities that have been fine-tuned and separated by strain. As a unique feature, AGORA2 collects microbial drug metabolism data that is broken down by strain. The microbiome and constraint-based modeling communities will be very interested in AGORA2, and it could be used for much more than that.
Article Source: Reference Paper | The 7,302 AGORA2 reconstructions are freely available at https://www.vmh.life/
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Sejal is a consulting scientific writing intern at CBIRT. She is an undergraduate student of the Department of Biotechnology at the Indian Institute of Technology, Kharagpur. She is an avid reader, and her logical and analytical skills are an asset to any research organization.