A team of researchers from the University of Edinburgh and collaborators from institutions like Stanford and UC San Francisco have tackled a central question in biotechnology: can we make protein expression predictable and routine? Their review, published in Trends in Biotechnology, explores why expressing proteins in host organisms remains a challenge, what’s holding the field back, and how we might finally “solve” protein expression.
Why Predictive Models for Protein Expression Matter
Protein expression sits at the heart of most scientific breakthroughs and industrial applications, powering everything from basic research to drug and enzyme production. Even so, pushing a brand-new protein into a fresh cellular host still feels like tossing darts blindfolded. That hit-or-miss work eats up funds, stretches schedules, and can put exciting projects on ice. The authors believe a smart, forward-looking model-not unlike the way AlphaFold revolutionized structure prediction-could shrink costs and kick reliability into high gear.
Factors Influencing Protein Expression
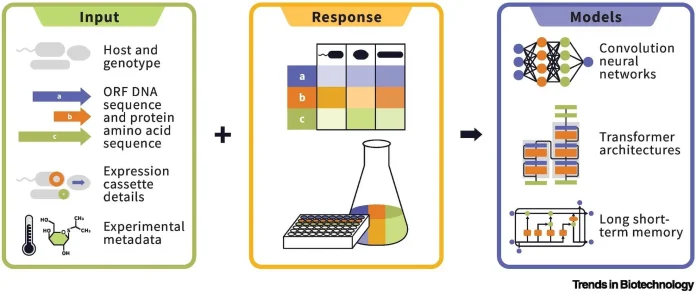
The complexity of protein expression comes from two main sources: intrinsic and extrinsic factors. Intrinsic factors are hardwired into the protein’s amino acid sequence. They determine whether the molecule folds, stays in solution, and holds up over time. Extrinsic conditions reflect every lab choice made by the scientist, the host cell, the promoter, recipe tweaks, and growth temperatures. Both sets of factors interact in complex ways, making it difficult to predict which combinations will yield a soluble, functional protein.
Current Limitations: Data and Models
After decades of work in the lab, scientists still lack a single model that reliably forecasts how much protein a gene will produce in any given host or setting. The data we have is usually tied to just one organism, generated with shifting protocols, or so small that it offers little to machine-learning tools. Most predictors zero in on narrow topics-codon choice, say, or solubility- and even these focus areas crumble under real-world tests. Take codon-usage apps: they are popular yet often pull different conclusions and never promise success on their own. The picture leaves experts calling loudly for larger, broader, and uniform datasets to build sturdy prediction tools.
The Need for Better Datasets
To move the field forward, the researchers urge the creation of a next-gen resource that is vast, varied, and freely shared. Such a collection would track expression for many proteins across multiple hosts, follow the same lab steps, and log both protein features and environmental factors. With that backbone, machine-learning models could learn not just whether a protein might be made, but also what settings to tweak so that it actually succeeds.
Roadmap for Data Collection and Model Building
The researchers outline a practical roadmap for generating this critical dataset. The team advises kicking off experiments with well-characterized microbes such as E. coli and Pichia pastoris, which are widely used, easy to grow, and can be scaled up easily. Data can then be harvested quickly through high-throughput pooled assays, whether label-free proteomics or growth screens, and those results should be double-checked against the classic single-sample workflows. Finally, everything goes into a clean, community-friendly format so future researchers- and their machine-learning tools-can grab and use the data without extra translation work.
Conclusion
Once the dataset exists, machine-learning models can comb through protein sequences and experiment notes, learning to predict which constructs will express well and which will flop. The researchers point out that protein language models-algorithms that sponge up millions of sequences much as ChatGPT digests text-are a natural fit for this task. With enough examples, such models may uncover the hidden grammar of protein expression, taking the field from educated guesses to precise, data-driven design.
Solving protein expression would have far-reaching impacts, enabling faster drug development, more efficient protein engineering, and deeper exploration of the protein universe. The authors acknowledge the challenges ahead, from data standardization to community coordination, but remain optimistic. By investing in systematic data collection and embracing machine learning, the field can finally move beyond trial and error and make protein expression as routine and reliable as DNA sequencing.
Article Source: Reference Paper
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.