Researchers at the University of Copenhagen, Denmark, introduce the Clinical Knowledge Graph (CKG), an open-source platform with about 20 million nodes and 220 million relationships representing relevant experimental data, public databases, and literature.
Evidence-based precision medicine has grown to include a more thorough examination of disease characteristics. This necessitates the seamless integration of a wide range of data, including clinical, laboratory, imaging, and multi-omics information. The necessity to collect, organize, and format relevant data has long been recognized in the biomedical research community, resulting in the widespread use of many biomedical databases. Mass spectrometry (MS)-based proteomics has come a long way in the previous decade; it now provides a more comprehensive picture of biological processes, cellular signaling events, and protein interactions.
Recently, a more fine-grained definition of disease that integrates clinical and molecular data has been discovered to give a deeper knowledge of individuals’ disease phenotypes and identify candidate markers of prognosis and/or treatment. As a result, systems that integrate numerous data types while capturing the links between molecular entities and the illness phenotype are required.
Knowledge Graph Concept
A knowledge graph or semantic network depicts the interaction between a network of real-world elements, such as objects, events, situations, or concepts. The name “knowledge graph” comes from the fact that this information is frequently kept in a graph database and represented as a graph structure.
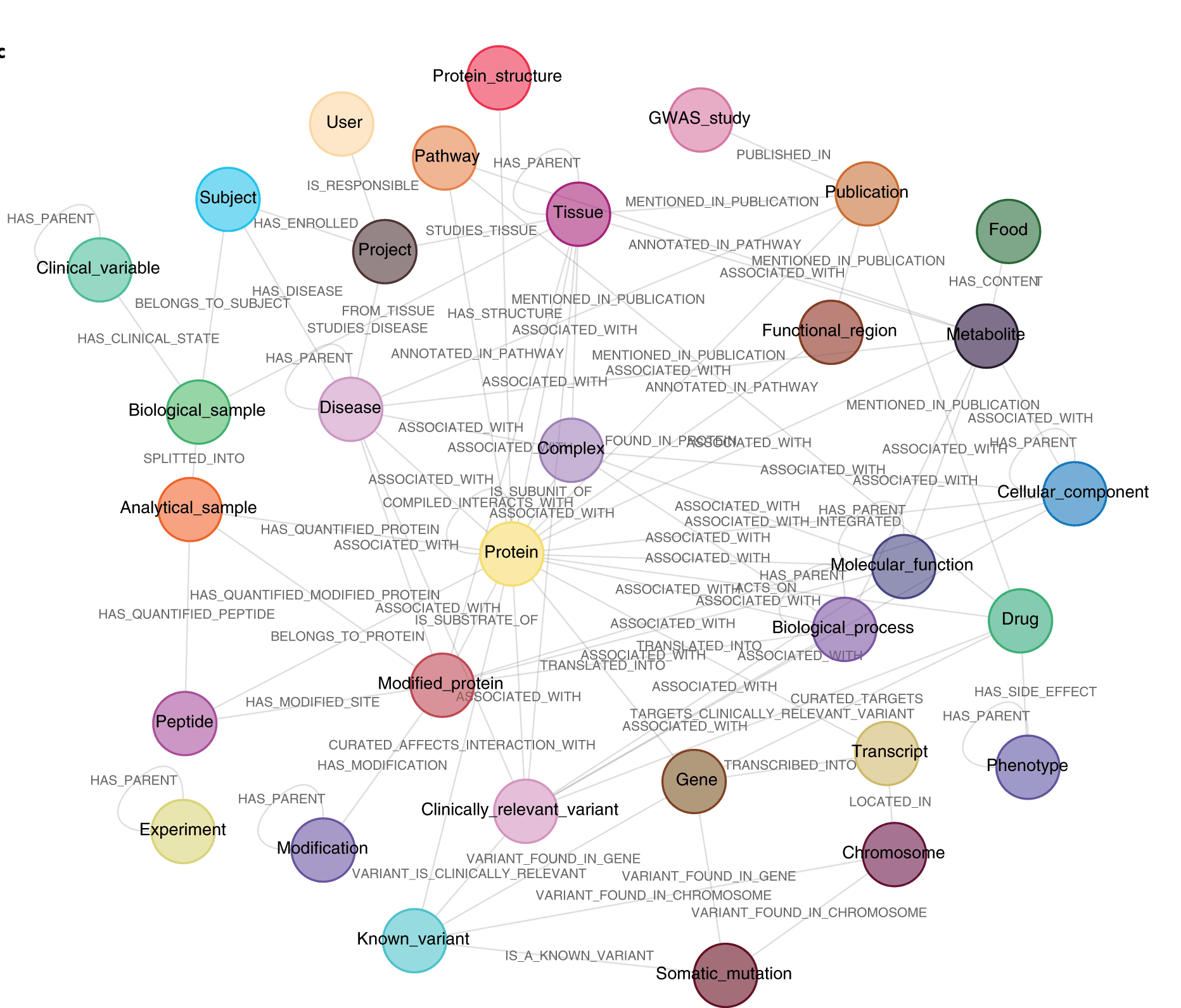
Image Source: A knowledge graph to interpret clinical proteomics data
The researchers offer a knowledge graph framework that allows for the integration of proteomics and other omics data while also including relevant biomedical databases and information taken from scientific articles. It is called the Clinical Knowledge Graph (CKG), and it’s a graph database with millions of nodes and relationships. It enables automated data processing, knowledge mining, and visualization, as well as clinically meaningful searches and advanced statistical analyses. By relying on scientific Python libraries, the CKG combines community work while making the platform dependable, maintainable, and ever-improving. The entire system is open-source and distributed under a permissive license. In both regular procedures and interactive exploration, it offers reliable, reproducible, and visible analysis.
The CKG is made up of several independent functional modules that: (1) format and analyze proteomics data (analytics_core); (2) build a graph database (graphdb_builder) by combining data from a variety of open access databases, user-conducted experiments, existing ontologies, and scientific publications (3) connect and query this graph database (graphdb_connector); and (4) enables data visualization and analysis with the help of online reports (report_manager).
The analytics_core of the CKG is built on an open modular design that is fully implemented in Python, using widely used and well-maintained open-source libraries that cover a wide range of data science topics such as statistics, network analysis, machine learning, and visualization. These libraries ensure the quality, robustness, and efficiency of the underlying algorithms and methods. It also allows for the introduction of new data science discoveries that can be readily modified to support proteomics data processing. The CKG also addresses growing worries about scientific results’ repeatability. Jupyter notebooks were used to create shareable analysis pipelines that make results repeatable and replicable, the researchers hope that this methodology will be widely adopted by the proteomics community and beyond.
The graphdb builder module loads the ontology, database, and experiment files into the graph database with a sequence of Cypher queries that produce the necessary nodes and relationships once they have been standardized, formatted, and imported.
The CKG database is constantly expanding, and it presently collects annotations from 26 biological datasets using ten ontologies and organises them into about 20 million nodes with 220 million links.
The network structure was discovered to be easily scalable, allowing new ontologies, databases, and experiments to be simply integrated. Furthermore, because of this intrinsic adaptability, nodes and associations that were initially created to give biological context for large-scale proteomics data interpretation can easily be remodeled to include other omics datasets.
The CKG’s primary purpose is to combine the analytics module’s power with the vast prior data stored in the graph database. The harmonization of numerous diverse but linked data sources enables standard analysis pipelines that produce results instantly, replacing weeks of human labor in a more consistent format. These basic reports give an initial assessment of the generated data’s quality, highlight relevant findings, and contextualize these hits in connection to the graph’s various biological components. The report manager manages the design and maintenance of experimental projects, as well as automated analysis, visualization, and knowledge extraction.
The “Data Upload” dashboard software integrates the clinical and/or proteomics data into the graph once it is ready and processed. All reports, analysis results, and visualizations can be downloaded as a single compressed file containing tables and figures in ready-to-publish format, in addition to being viewable in the browser.
CKG’s default pipeline was utilized in proteomics research of nonalcoholic fatty liver disease to demonstrate how it speeds up and extends data analysis and interpretation. The automated analysis pipeline represented all clinical, proteomics, and multi-omics analyses as a graph, with all regulated proteins and relationships extracted from the knowledge graph (such as diseases, drugs, interactions, and pathways) prioritized and reduced the number of nodes presented using betweenness centrality. The default pipeline took less than 5 minutes to complete, yet it caught almost all of the insights gained from our previous manual study, which had taken weeks. Time-consuming literature and database searching for known/published protein–disease connections, as well as knowledge gathering, had gone into the interpretation of the differentially abundant proteins. Still, the CKG revealed them to be incomplete.
The researchers reanalyzed a recent study in which they discovered cancer/testis antigen family 45 (CT45) as a biomarker for long-term survival in serous ovarian adenocarcinoma and detailed its method of action to test the CKG’s multi-analysis capabilities. CT45 was much higher expressed in patients with long-term remission after chemotherapy, according to the CKG. The CKG also verified that little was known about CT45’s cellular responsibilities and functions before. However, it did produce 24 putative CT45 interactors, four of which belonged to the PP4 complex and were contributed by a human interaction map. The CKG identified numerous known DNA damage kinases and their regulated substrates, shedding light on carboplatin’s mechanisms of action and the negative regulation of proliferation via DNA damage repair. In addition, the CKG revealed a number of other important kinases and relationships that had previously evaded manual analysis.
“After standard treatment options have been exhausted in end-stage cancer, molecular profiling might still reveal druggable targets and opportunities for drug repurposing, and previously proteomics profiling of cancer tissue was used to identify alternative targeted strategies.” On this idea, CKG is used to prioritize treatment options for chemorefractory cases and currently, it mines more than 350,000 connections between proteins and approved or investigational drugs targeting them.
The CKG’s open nature and free availability may enable the accumulation of data and knowledge into a community graph. This would ensure that similar proteomics or omics experiments carried out elsewhere benefit the community. This extends the ‘rectangular strategy’ for biomarker discovery, allowing direct and profound project comparison and resulting in progressively robust and powerful analysis and information generation.
According to the researchers, different groups and institutions will have their own local version of the CKG, which will secure the sensitive nature of healthcare data while yet allowing cross-platform studies. New approaches, such as differential privacy and federated learning, would allow other researchers to use the CKG to train models iteratively across institutions without direct access to sensitive data. Artificial intelligence is set to play an increasing role in MS-based proteomics and biomarker discovery, and the researchers look forward to integrating the CKG with these capabilities as well as taking advantage of novel graph deep learning capabilities.
Story Source:
Santos, A., Colaço, A.R., Nielsen, A.B. et al. A knowledge graph to interpret clinical proteomics data. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-021-01145-6
Data Availability: https://doi.org/10.1038/s41587-021-01145-6
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Swati Yadav is a consulting intern at CBIRT. She is currently pursuing her Master's from Delhi Technological University (DTU), Delhi. She is a technology enthusiast and has a keen interest in the latest research on bioinformatics and biotechnology. She is passionate about exploring new advancements in biosciences and their real-life application.