Researchers from the Center for Computational Life Sciences at the Cleveland Clinic introduce R2G2, a Python-R framework that automates the integration of R and Bioconductor tools into the Galaxy platform. As a bioinformatician who lives daily in RStudio, terminals, and Galaxy histories, this feels like someone just removed a chronic, low-grade headache from workflow development.
The problem: too many tools, too little time
R and Bioconductor have, over the years, become essential for computational biology, supporting over 2,289 software packages and several hundred annotation and experimental data packages, as of the 3.20 release. Coupled with the ever-growing ecosystem of Python (614,000 packages on PyPI as of early 2025), the community has access to countless tools to analyze the various omic data types. However, what the community is lacking is the transition of tools and methods from development to implementation. This is particularly true for wet-lab researchers, who require the bioinformatics tools but do not code.
Surveys show that researchers in the life sciences use bioinformatics tools, but also show that more than 50% of researchers report little to no bioinformatics knowledge, 74% report no programming knowledge, and the majority report they have little statistical knowledge. Bioinformatics platforms like Galaxy have tried to bridge the divide by encapsulating low and high-level programming with a workflow management system that is executable from a web interface and can be shared and versioned. However, there are still only 82 Bioconductor packages integrated with Galaxy, which include some of the more popular ones like limma and DESeq2, leaving a plethora of R packages that require scripting to access. The blocker is not willingness, but the manual effort required to create and maintain Galaxy XML wrappers, wire dependencies, test tools, and keep everything in sync as packages evolve.
How R2G2 bridges R and Galaxy
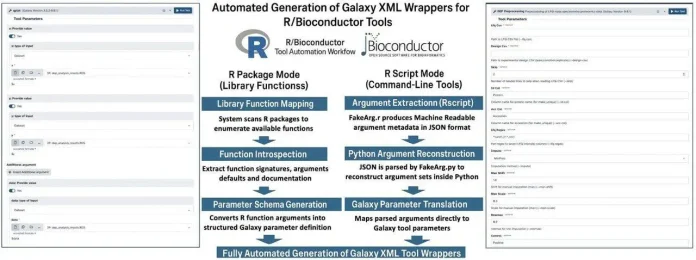
R2G2 aims to ease the tedious work between R and Galaxy. It has two main modes. One is generating Galaxy wrappers from R or Bioconductor packages. The other is from R scripts with argparse-style command line interfaces. For packages, R2G2 accesses an R package using the rpy2 interface, which allows it to analyze the package’s functions, get formal arguments, and retrieve help documentation from the associated .Rd files. With this information, R2G2 programmatically builds Galaxy XML wrappers for the functions, where R argument types (character, numeric, integer, logical, choices, and file) will be mapped to Galaxy parameters (text field, number field, boolean, and dropdown).
For each wrapped function, R2G2 generates an R script to be run in Galaxy. This R script will load the necessary package, convert the Galaxy inputs to R objects, call the target function, and write the outputs in RDS format, which is compatible with downstream tools. It even documents and codifies flexible R constructs like ellipsis (…) parameter to be translated to repeatable and conditional inputs in the Galaxy interface. In the script-based mode, the authors implement a FakeArgs class in R (using R6 and r-argparse) that pulls argument definitions from a script and exports them to JSON. R2G2 can also take build tools JSON and convert it to the Python-style argument descriptions using CustomFakeArg (a class from the anvi’o ecosystem), rebuilding groups and conditionals, and then constructs everything into a Jinja2 template to produce a complete and usable Galaxy tool.
The framework fits well into the existing Galaxy ecosystem. Dependencies are resolved via Conda (also mamba, Docker, or Singularity), so correct versions of R, Bioconductor, and related packages will be available at execution time. Tool authors can use Galaxy development and publishing workflows to test their tools before pushing them to the Galaxy ToolShed, where the rest of the Galaxy community can access them.
Real-world demonstrations
The authors show R2G2 in action via some bioinformatics workflows. One such workflow is the in-house pipeline based on the Bioconductor DEP package for label-free quantitative proteomics. With R2G2, they can turn scripts like DEP_preprocessing.r into Galaxy tools that accept MaxQuant protein tables and experimental design files and perform filtering, normalization, and multiple imputation (MinProb, MinDet, kNN, QRILC, etc.) and generate processed data tables and quality control PDFs. Another tool within Galaxy incorporates the script DEP_DE_analysis.r, which allows users to conduct differential expression analysis, set their own thresholds for P-value and log2 fold-change, and create PCA and volcano plots, all of which can be customized through Galaxy.
To illustrate the generality of their tool, they use R2G2 on 41 R scripts, available in different repositories, that the authors describe as performing INDEL detection (indelfindr.R), identifying CpG islands, splitting and trimming FASTA sequences, and processing PDBs. These command-line scripts are turned into Galaxy tools that need no more than a couple of minutes of operator effort. The authors also looked through the Bioconductor library for scripts that use argparse and found 51 such scripts from CircSeqAlignTk, MAGAR, RnBeads, infercnv, and openCyto, all of which were successfully converted to Galaxy. In what is perhaps their most striking experiment, they apply R2G2 to ggplot2 and obtain more than 450 Galaxy tools, created at the function level, indicating a potential for more granular and easily manipulated interface (GUI) components for building visualizations.
Why it matters
Unlike other versions of R to Galaxy, R2G2 literally changes who can realistically bring methods into Galaxy. Instead of expecting every R developer to become an expert on Galaxy’s XML schema and deployment tooling, R2G2 empowers them to write clean packages or well-structured scripts and rely on automation for integration. For wet-lab scientists and trainees, containerization and versioned environments promise reproducibility and faster access to a much larger slice of the Bioconductor universe, all within an interface they trust.
The authors of R2G2 openly acknowledge the tool as not being a perfect one-click solution; output inference can require manual checking, large packages can produce a tool set that is unmanageable, and expanded automated testing would strengthen confidence further. Nevertheless, R2G2 v0.1.1 has been released as a PyPI package, and the authors have shared their generated wrappers and reference scripts on GitHub, providing a pragmatic start that labs can adopt and extend, as their bioinformaticians wrestle with XML wrappers. For many, the real impact is subtle but important: the bioconductor tools are there when needed, and more time can be used on study design, interpretation, and collaboration.
Article Source: Reference Paper | Availability: R2G2 is available as a PyPI package at https://pypi.org/project/r2g2/0.1.1/. Its source code can be downloaded directly from GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.















