When researchers at the Department of Biology and Biotechnology, the University of Pavia, set out to rethink bioinformatics teaching. They started from a simple observation: students are getting very good at running tools, but not necessarily at telling the biological story behind the output. Their new pipeline, “Eduomics”, is an attempt to fix exactly that gap by turning next-generation sequencing simulations into complete clinical narratives that students can explore, question, and interpret.
Why bioinformatics teaching needs an upgrade
As the costs of sequencing drop and the use of multi-omic data becomes more widespread in industry and academia, bioinformatics moves from a niche skill to a fundamental form of literacy in the life sciences. And yet, a large part of current training is still what some have termed “buttonology.” Learning what command to run, what parameter to change, what file format to use, and which tool to use.
There are no ethical or privacy issues with using simulated datasets, yet most existing simulators are designed for benchmarking, not for teaching purposes. Educators are presented with a jumble of unconnected tools, each with its own dependencies, input formats, and configuration options, and have to manually integrate the simulated data with a biological or clinical context. This leads to a common outcome: the simulations are underutilized, and to the extent they are not, students can complete an entire analysis at the end of which they do not have an appreciation for “who the patient is” behind the VCF or the count matrix.
What does eduomics actually do?
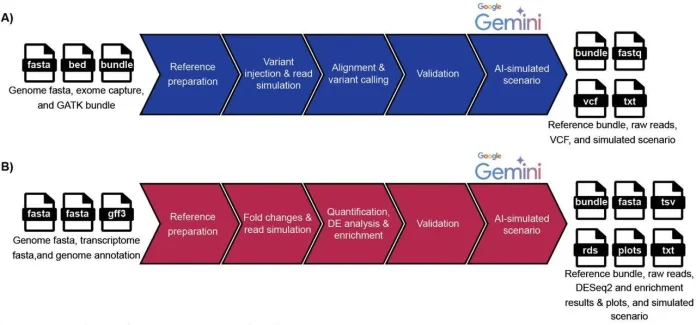
The eduomics pipeline has been developed using the Nextflow DSL2 language, combining modular processes in line with nf-core community standards. It finished all simulations, advancing through major NGS use cases: variant calling and RNA-seq. It only requires the educator to fill out a CSV sample sheet with data on chromosome, type of workflow, coverage, and number of replicates.
From there, the pipeline takes over. For DNA resequencing, eduomics injects known likely pathogenic and pathogenic ClinVar variants into a selected genomic interval, simulates case–control reads, and runs them through a GATK Best Practices–style workflow. In the case of RNA-seq, it first creates expression patterns by vertically integrating co-regulated gene clusters with the help of Gene Ontology, simulating reads, and then quantifying them. In the second stage, differential expression is measured with an analysis performed with Salmon and DESeq2. Importantly, in both branches, only simulations that reproduce the expected ground truth after analysis are kept, so Instructors know the data behaves in a controlled and interpretable way.
Storyline-based learning: beyond problem sets
Eduomics attributes particular importance to the events that follow the completion of the bioinformatics component of the project. Here, the validated sets of variants and lists of differentially expressed genes are transferred to a specialized unit that uses the Gemini API to produce elaborate, patient-inspired clinical stories. These narratives include symptoms, family history, biochemical findings, imaging findings, and possible diagnoses consistent with the underlying molecular changes.
This design incorporates what the authors term “storyline-based learning”. The students are not just solving a technical task (like “call variants on these FASTQs”), but intertwining a sequence of technical and clinical events. The students, for example, encounter a simulation that describes a stop-gain mutation in the PEX26 gene on the 22nd chromosome, while eduomics presents the clinical picture of a child with hypotonia, developmental delay, retinal dystrophy, and elevated levels of very long-chain fatty acids, which is a real description of a peroxisome biogenesis disorder. The students also understand, in a stepwise manner, how a systemic disease that is complex can be caused by a single mutation that is found in a membrane protein of a peroxisome.
Under the hood: realism and scalability
Despite being framed educationally, the work being done around chromosome bundle construction, partitioning, and the integration of various public resources, including gnomAD, ClinVar, AWS iGenomes, and simulation tools like SimuSCoP for realistic Illumina read generation, is serious technical heavy lifting. While capturing regions, the work on the educational RNA-seq side draws on GO term networks and community detection to delineate gene sets likely to yield meaningful enriching results, avoiding spurious patterns of DE that have no productive biological consequence.
The authors tested the pipeline on human chromosome 22, operating on both a local HPC and a Google Cloud instance. In one example, the workflow took almost seven hours to run, completed around 12,000 jobs, and cost approximately €199, a clear and convincing proof of the simulation’s successful scalability and portability. With about 38 percent of the injected pathogenic variants recovered within the DNA workflow, and almost 79 percent of the RNA-seq simulations meeting the criteria of successful DE and enrichment, the results confirmed both the feasibility of the pipeline and the complexity and robustness of the workflow.
Conclusion: What does this mean for teachers and students?
Eduomics makes it easier to use heavy simulations in workshops, courses, and large online courses. One configuration file can create controlled, unique datasets and clinical stories. These datasets can be used to create unique cases for student use. Each student or group can be given a case that is personalized and is still grounded in a known truth. This is very useful for assignments, where tutors assess students based on cases. The pipeline is built on Nextflow, which is modular, allowing it to be used on different computational infrastructures, including HCP and cloud, a useful feature for institutions that have varied computational resources.
For students, the payoff is cognitive rather than just technical. Rather than just “Did the pipeline run?”, students ask “Does the result make sense for the patient?” These shifts provide the students a chance to form and test a hypothesis, integrating and connecting GO terms and pathways to varying phenotypes. The students learn to see NGS info as part of a larger clinical and biological context instead of just examining the data in isolation. This education empowers them to do more than just use the tools and to think like real scientists and bioinformaticians in the context of biomedicine.
Article Source: Reference Paper | Website | GitHub
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.