Scientists from BGI-Research developed a new version of the Cultivated Genome Reference (CGR), a repository of high-quality draft genomes of the human gut microbiome. The current version of CGR, which is CGR2, has been further expanded to incorporate numerous high-quality draft genomes generated from cultivated bacteria. CGR2 classifies previously unidentified species and uncovers the functional and genomic diversity of bacterial strains. An in-depth analysis of carbohydrate-active enzymes (CAzymes) reveals the phyla with the largest and most diverse repertoires of these enzymes. CGR2 also enabled the identification of genes involved in the synthesis of secondary metabolites in the gut microbiome. The unraveling of the gut microbiome genomic landscape will enable the development of therapeutics and provide a deep insight into the evolution of the human gut microbiome.
CGR and other precursors to CGR2
Over the last decade, it has become increasingly clear to the scientific community that there is an underlying association between the gut microbiome and metabolic traits and diseases. Fecal transplantation has significantly aided disease management, and in the future, customized transplantation with a well-characterized microbiome will be desirable for clinical interventions. This requires an in-depth classification and analysis of the human gut microbiota.
Culture-independent techniques have enabled the discovery of several uncultivated organisms in the gut. The Unified Human Gastrointestinal Genome (UHGG) repository has an unprecedented number of bacterial genomes pertaining to the human gut. However, a significant percentage of these species lack cultivated isolates.
Cultivation-based studies have provided deeper insights into the study of the gut microbiome. In the previous catalog of reference genomes of the human gut microbiome, CGR, the authors presented a reference catalog with improved taxonomic annotation and functional inferences. Cultivation-based resources enable better identification and isolation of carbohydrate-binding enzymes, bacteriophages, and probiotics. Thus, the authors further expanded the CGR repository and developed an expansive repertoire of human gut microbiome draft genomes, CGR2.
CGR2 has been developed using a large-scale culture-based method for obtaining reference human gut microbiome genomes and enables a more expansive exploration of the human gut microbiome, thus establishing CGR2 as a valuable resource for gut microbiome research.
CGR2 unravels the genomic landscape of reference human gut microbiome genomes
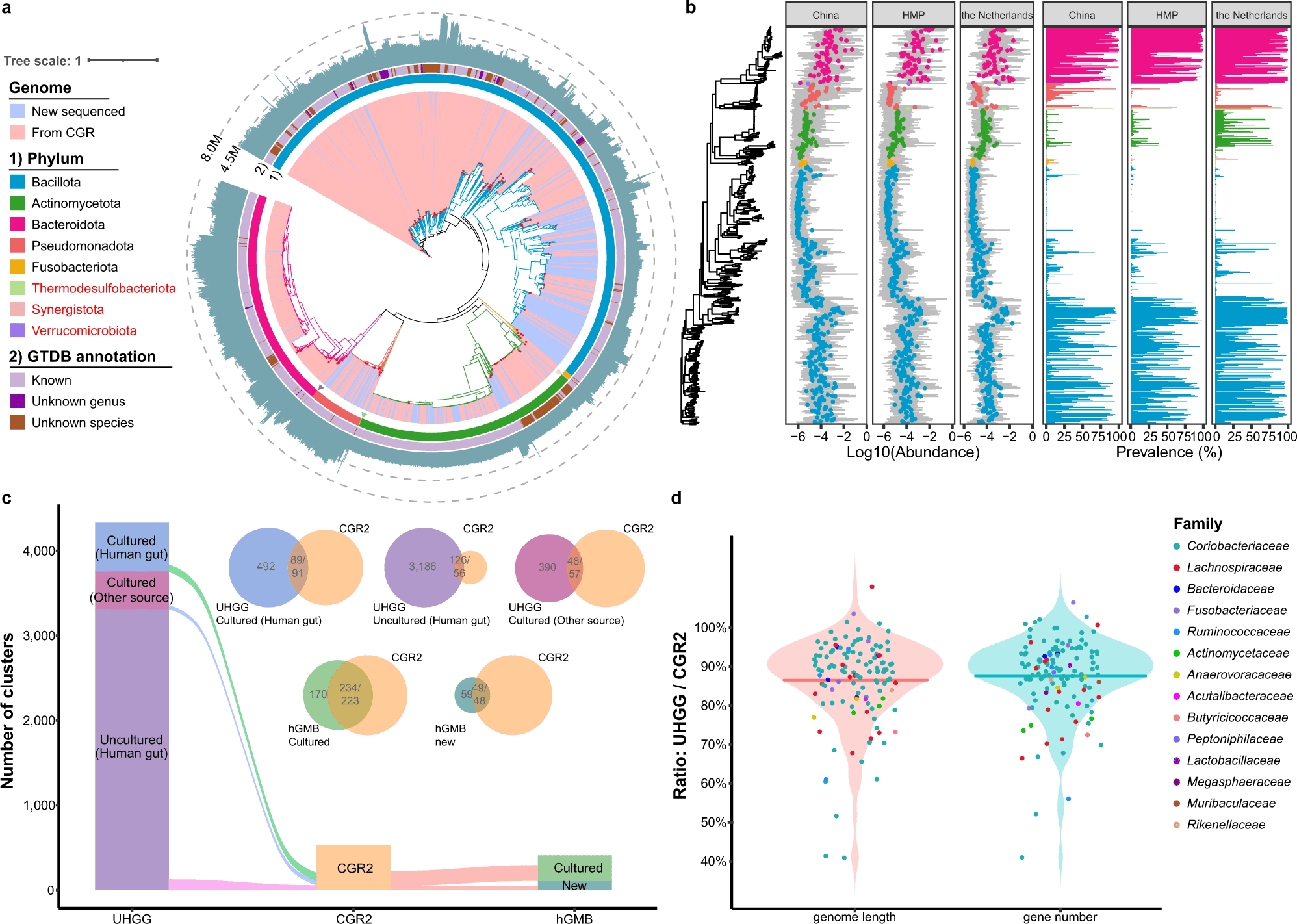
CGR2 provides an expanded repertoire of gut microbiome genomes. The previous version, CGR, presented more than 1500 reference genomes for the human gut microbiome. The current iteration, CGR2, provides 3,324 high-quality genomes clustered into 527 species-level clusters based on 95% average nucleotide identity (ANI). Of these, 179 were not classified at the species level and thus represented previously unidentified species.
Next, the authors performed an in-depth analysis of CAzymes (carbohydrate-active enzymes). This revealed the function of the culture isolates in CGR2 and generated the distribution of CAzymes across different phyla. Isolates belonging to the phylum Bacteriodota revealed the largest and most significantly diverse repertoire of CAzymes.
CGR2 also enabled the identification of genes involved in the synthesis of secondary metabolites. A comprehensive analysis of secondary metabolite biosynthetic gene clusters (SMBGs) was performed using AntiSMASH. More than 4000 gene clusters were identified in more than 2000 genomes. Phylum Bacillota harbored the highest abundance of SMBGs.
CGR2 also enabled the prediction of prophages in the phage-bacteria interactions in the gut microbiome. To obtain detailed networks between phages and the host bacterium, the authors used VirSorter. A phylogenetic tree depicting the evolutionary characteristics of gut phages was constructed.
The following figure illustrates the phylogenetic tree.

Image source: https://doi.org/10.1038/s41467-023-37396-x
The authors performed a genome-wide analysis for Collinsella aerofaciens, which revealed high genomic and functional variation. More than 10 clusters were identified in CGR2, pointing towards high diversity amongst the bacterial clade.
The following figure illustrates the taxonomic profile of CGR2.

Image source: https://doi.org/10.1038/s41467-023-37396-x
Conclusion
The authors developed a large-scale cultivation-based repository for reference genomes of the human gut microbiome. This is indeed a comprehensive and valuable resource as it provides deep insights into the genomic landscape and functional diversity of the gut microbiome. The role of the gut microbiome in several diseases as well as in metabolism has been well established, and it is necessary to gain a deeper understanding of the diversity and characteristics of the various organisms present as part of the microbiome. Owing to the rising antibiotic resistance worldwide, a better insight into phage-host bacterium interactions can aid in the development of anti-resistance treatment against bacterial infections. The identification of previously unidentified species and the CAzyme analysis reveals significant insights into the microbiome interactions that will further aid in the development of next-generation probiotics. The biomolecules produced by the microbiome are significantly crucial in the agricultural as well as pharmaceutical sectors. Such knowledge will also enable customized fecal transplantation in the future, which will drastically improve therapeutics for several gastrointestinal and gut-related diseases and provide better methods for clinical interventions. Thus, CGR2 is an impressive expanded version of the previous CGR, adored with multiple capabilities of a valuable resource for the human gut microbiome research community as well as the society at large. The authors also envision that such insights will aid in the development of secondary metabolites, thereby presenting alternative therapeutics.
Article Source: Reference Paper
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.