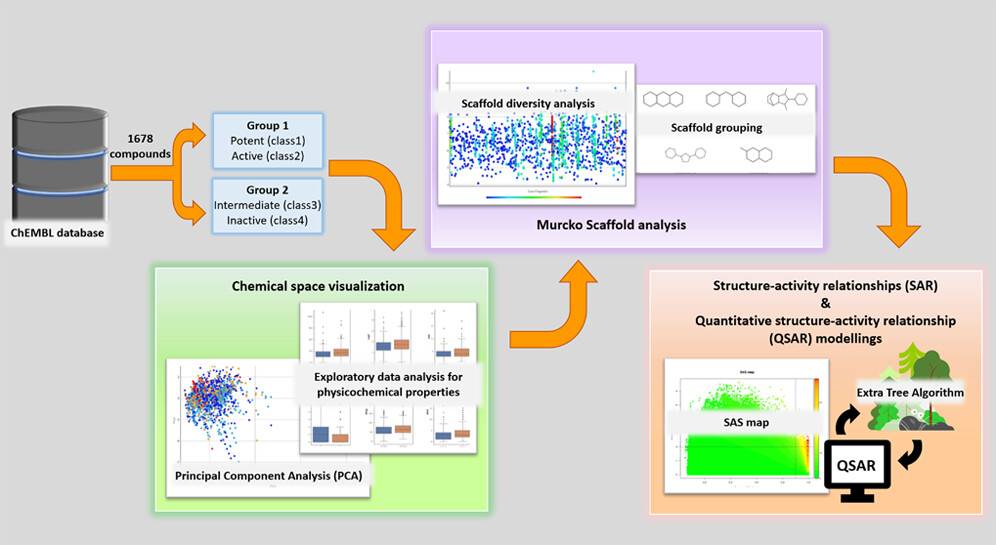
Researchers from Thailand perform a systematic cheminformatics analysis and machine learning modeling to study how human Androgen Receptors (AR) antagonists combat Prostate cancer, one of the leading causes of death in male cancer patients. Androgen receptor signaling drives the growth and prostate cancer progression. The authors perform the study on human AR antagonists from the ChEMBL database to visualize the chemical space and analyze the structure-activity relationship and landscape, as well as the Murcko scaffold. These findings can guide future attempts at drug discovery for combating prostate cancer.
Androgen Receptor (AR) signaling and AR antagonists
Prostate cancer is one of the leading causes of death in male cancer patients. The mechanisms underlying the pathogenesis and progression of Prostate cancer are primarily associated with androgen synthesis and androgen receptor signaling pathways. It has been seen that for localized advanced cases of the disease, androgen deprivation therapy (ADT) through medication or surgery can slow down the progression of the disease. Thus, androgens are a crucial factor in the fight against prostate cancer.
The androgen receptor (AR) is a steroid receptor that belongs to the nuclear receptor superfamily. AR is involved in regulating the growth and development of the prostate by acting as a transcriptor factor. It has been found to be the pivotal regulator for disease progression and pathogenesis in prostate cancer and, thus, a significant therapeutic target for drug discovery against prostate cancer. It has also been found that AR is overexpressed in most prostate cancer cases and drives the disease progression to castration-resistant prostate cancer (CRPC).
AR antagonist development for the treatment of prostate cancer, especially CRPC, has been the prime focus of many studies. AR antagonists are of two kinds, viz, steroidal and nonsteroidal. The steroidal antagonists have been associated with undesired side effects and hence have been replaced by the nonsteroidal ones. These are the first generation of nonsteroidal antagonists as well as the second generation. The first-generation ones were found to induce mutations along AR’s ligand binding domain (LBD), which leads to partial agonism to the mutated AR. Hence, they were replaced with second-generation antagonists. Though an improvement over the first generation, the second-generation antagonists are found to have induced AR-resistant mutations rendering them partial or mixed agonists for AR. Hence, these challenges impose the need for the development of novel AR antagonists.
AR antagonist fighting prostate cancer: through the Cheminformatics lens
The multidisciplinary field of cheminformatics applies information and computation technologies to find solutions to a plethora of problems in chemistry. Drug discovery has benefited in manifolds from cheminformatics approaches applied to the search for novel molecules as well as their optimization. Needless today, the exponential growth in ML and AI-based methods have greatly advanced the field of cheminformatics. The authors of the current study conduct systematic cheminformatic analysis as well as machine learning modeling for facilitating the drug discovery of novel AR antagonists.
Cheminformatics approaches were applied for chemical space visualization and Murcko scaffold analysis.
Chemical Space Visualization: All molecules are grouped under group 1 (potent/active classes) and group 2 ( intermediate and inactive classes). Based on the calculation of the six physiochemical properties of the groups, they are visualized and compared with each other. The following figure illustrates the chemical space visualization for AR antagonists using Principal Component Analysis (PCA).
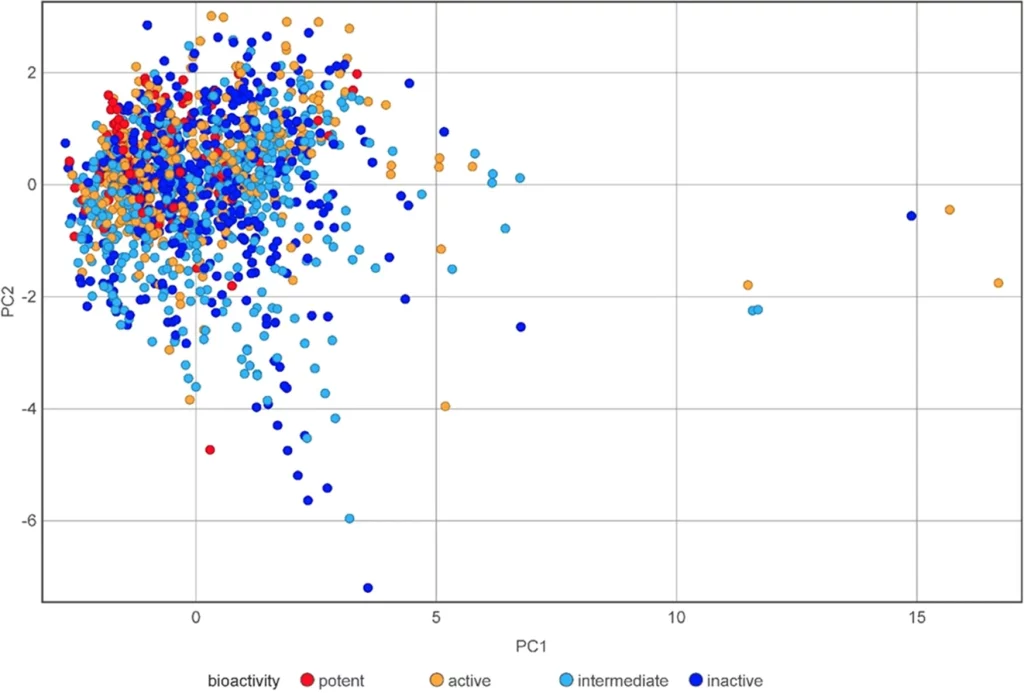
Image source: https://doi.org/10.1021/acsomega.2c07346
Murcko scaffold analysis: Scaffold analysis involves three components, viz. scaffold visualization, scaffold diversity analysis, and scaffold correlation with bioactivities. The diversity analysis has revealed that the diversity of group 1 molecules is lower than the intermediate and inactive classes. This clearly signifies the need for more novel scaffolds for AR antagonists.
Machine learning modeling reveals the structure-activity landscape
QSAR, a quantitative structure-activity relationship, is a mathematical model that is used to correlate molecular structures to their bioactivities. The SAS map, the structure-activity map, is applied to the data, which is PubChem fingerprint information here, and QSAR is executed to reveal the structure-activity landscape for the AR antagonists.
Comparison with previous studies
A previous study conducted by Hao et al. has applied chemoinformatic analysis to AR agonists as well as antagonists using data from PubChem. Another previous study conducted by Ban et al. applied structure-based drug discovery approaches for the computational drug discovery of AR antagonists. The machine learning-based project CoMPARA is a collaborative modeling project for AR activity. In comparison to these previous studies, the authors have used data from the ChEMBL database and applied a ligand-based drug discovery approach.
Conclusion
Androgen receptor signaling plays a crucial role in the development as well as disease progression of prostate cancer. CRPC has also been associated with an overexpression of AR, which acts as a transcriptor factor and thus is a crucial target for therapeutics and drug discovery associated with prostate cancer treatments. In this research article, the authors have extensively studied and performed a systematic chemoinformatics-based analysis of the human AR antagonists from the ChEMBL database. These analyses led to the chemical space visualization, and the Murcko scaffold analysis identified 16 representative Murcko scaffolds and revealed that active/potent class molecules are sparse compared to the inactive/intermediated classes. This alludes to the need for the drug discovery of novel AR antagonists. The authors have also resorted to machine learning-based modeling, QSAR, for determining the structure-activity landscape for the AR antagonists. However, the study has a major limitation in terms of the fact that valuable datasets could be missing from ChEMBL as ongoing assays and experiments are not reported thereat. Nevertheless, the study points to a methodology direction that will facilitate future drug discovery for the treatment of prostate cancer.
Article Source: Reference Paper
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.