Scientists from China have recently performed pan-3D genome analysis for soybean genomes to investigate the interplay among 3D genomic variations, structural variations (SV) as well as gene expression. Researchers applied Hi-C techniques to 27 soybean genome accessions to provide a pan-3D genome landscape of soybean and shed light on the evolutionary dynamics of 3D genomes among plant accessions. The study reveals that compartment divergence is largely contributed by intersection regions between A/B compartments. Topologically associating domains (TADs) are also seen to exhibit varying densities across compartments. The study also shows how non-long terminal repeat retrotransposons (non-LTR), Gypsy elements, and satellite repeats influence TAD boundaries. The pan-3D genome analysis presents the seldom-reported diversity of the genome across plant accessions, and modifying the 3D genome structure can lead to future solutions for plant breeding.
Pan-3D genome analysis
DNA, along with histone proteins, is packaged into chromosomes in eukaryotic cells. The three-dimensional chromosomal structural organization brings distal cis-regulatory elements (CREs) into close proximity to their targets, thereby aiding gene expression and regulation. The 3Dimensional (3D) genome in eukaryotic cells is hierarchically organized into chromosome territories, compartments, topologically associating domains (TADs), and chromatin loops. Technological advances in microscopy and chromosome conformation capture methods, coupled with next-generation sequencing techniques, have revealed the complex 3D genome organization. Variations in the 3D genome organization exist between cell types as well as during the developmental stages of cells. While the mammalian genome organization analysis using techniques such as these has provided insights into the organization’s role, not much is known about the chromatin organization in plants.
The compartments are nonrandom yet dynamic structures. The transcriptionally active A compartments are characterized by long stretches of megabases interspersed with the B compartments that are transcriptionally inactive. TADs are contiguous genomic sequences known to exhibit higher interaction frequencies than those outside the TAD region. They are separated by TAD boundaries which can restrict CRE interactions with promoters in the TAD region.
Comparative genomic analyses have revealed that changes in the chromatin structure can drive processes from cell differentiation to embryo development and even genomic evolution. It has been reported that genomic structural variations (SVs) can change the arrangement of the 3D genome, thereby affecting gene expression for several genes.
While most previous studies involving plant genomes have dealt with genomic sequence information, proving valuable and critical biological insights, studies aimed at understanding the 3D genomic landscape among plant accessions are limited. Furthermore, due to a lack of high-quality SV data, the influence of SVs on the 3D genome has not been fully characterized, especially for plants.
The authors performed pan-analyses on the 27 de novo (previously conducted genome assembly) soybean genome accessions and constructed the pan-3D genome of A/B compartments and TAD boundaries.
Methods employed for constructing the pan-3D genome
The authors employed in situ Hi-C sequencing of the 27 accessions of soybean that included wild, cultivars and landraces variety of soybean. A robust contact matrix resolution of 25 kb resulted in a saturated 3D chromatin structure.
Outcomes of the pan-3D genome analyses
- Identification of A/B accessions revealed that while overall compartment percentages are conserved, reorganization of A/B compartments within chromosomes is frequent in soybean accessions.
- The variation of A/Bcompartmets is closely related to genomic features.
- Compartment switching was observed in regions with intermediate genomic features.
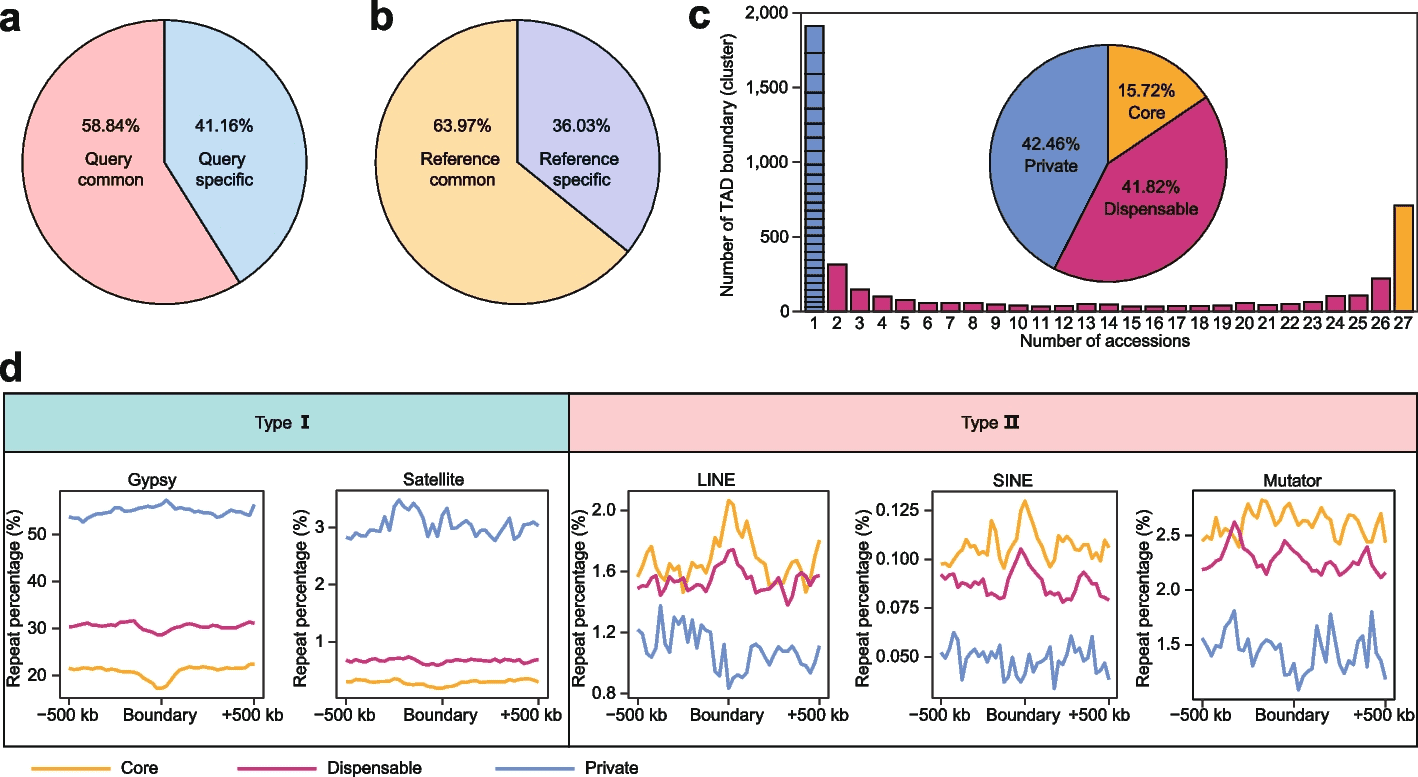
- The pan-3D genome construction for TAD boundaries revealed that TAD boundaries have a higher level of variation than A/B compartments.
- Also,non-LTRs were enriched around TAD boundaries, alluding to their importance in maintaining TAD boundaries.
- Satellite repeats and Gypsy elements were also found to be enriched around the TAD boundaries. The authors were the first to show this.
- Analyses to study the interplay between genomic SV and 3D genomic variation revealed that Presence and Absence Variations(PAVs) play a crucial role in 3D genomic variation.
The following figure illustrates the pan-3D genome for the TAD boundaries.
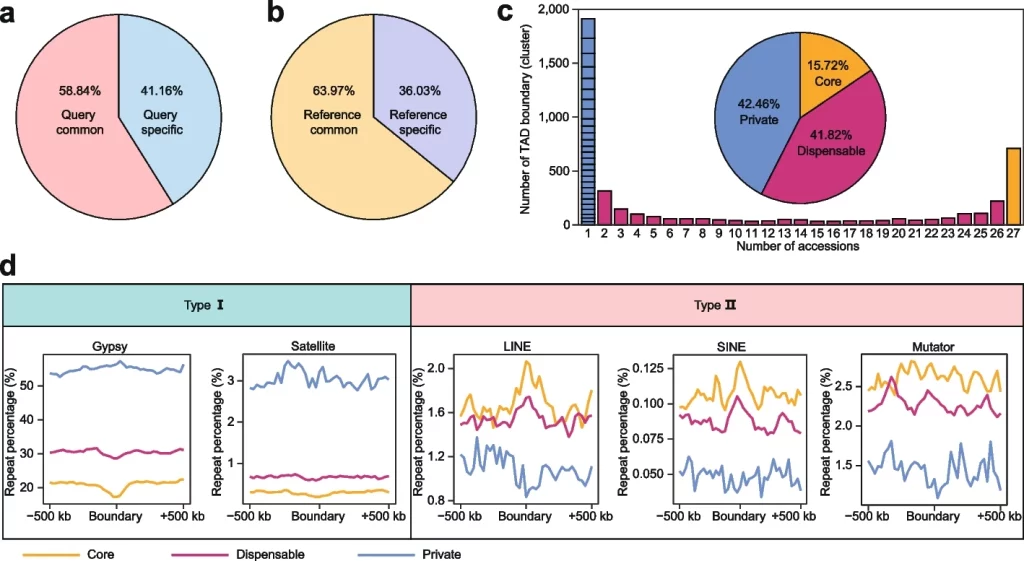
Image source: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-023-02854-8/figures/4
Conclusion
The authors have constructed pan-3D genome landscapes for 27 soybean accessions which have resulted in novel findings in terms of plant genetic diversity. The association between genomic SVs and 3D genome variation has been clearly established, as we see in the outcomes. This study is the first to report that transposable elements reshape the 3D genome in plants. The TAD boundary pan-3D genome constructions revealed remarkable insights into the role of several genetic elements in shaping the three-dimensional chromatin structure. This is also significant for understanding genome evolution in plants and also for implementing future solutions in crop breeding.
Article Source: Reference Paper 1 | Reference Paper 2 | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.