Scientists from Amsterdam, Netherlands, have developed an automated and open-source workflow for the design of 3D fragment libraries in the KNIME software. Fragment-based drug discovery (FBDD) has proven successful in producing a growing list of clinical candidates, with six approved drugs as of 2022. Fragments, or smaller molecules, are used as screening compounds as opposed to high-throughput screening (HTS) techniques. These small molecules typically have low affinity and activity for drug targets and hence require more sensitive screening techniques as opposed to biochemical assays for detecting binding, for example. The current workflow ensures the diversity in terms of physiochemical properties, molecular shape, and fingerprints of the novel, three-dimensional fragments selected as library members. The authors demonstrate workflow usability in FBDD by designing a focused 10-member fragment library based on the cyclopropane scaffold.
What is Fragment-based Drug Discovery?
Fragments are small molecules that are used as screening compounds in the drug discovery process for the FBDD approach. Compared to the HTS approach, FBDD involves designing fragment libraries that are required to comply with the empirical guidelines of the Rule of Three (Ro3), which define ranges of physiochemical properties for fragments. Fragments, being small molecules, have very low affinity and activity for drug targets and require more sensitive screening techniques than routine biochemical assays. Fragment libraries typically have a few thousand screening entities, and the design process should thus incorporate chemical diversity and appropriate physiochemical properties. It is also necessary to avoid substructures with adverse properties such as assay interference, aggregation, and reactivity. Another crucial point to keep in mind while designing fragment libraries is fragment sociability, which is the extent to which a screening hit lends itself to amenable chemistries, allowing a quick and efficient hit to lead optimization.
The Library Designing process and fragment library designing
In recent times, the design and development of chemistries and libraries of 3D molecules have received increasing effort and attention. It has been widely accepted that the fraction of sp3 -hybridized carbon atoms (Fsp3 ) in drug candidates highly correlates with clinical success rates as well as improved solubility and reduced promiscuity. Other metrics implied to assess the three-dimensionality of compound libraries are the principal moment of inertia (POI) and the plane of best fit (PBF). While most current fragment libraries largely have flat molecules, incorporating the 3D characteristic into the screening fragments would lead to the generation of leads and clinical candidates with such 3D characteristics, which are expected to show an increase in the novelty and diversity of the resulting hit compound.
Guidelines for the Fragment Library Design
A host of guiding tools exist for guiding the process of library design. Various computer-aided drug design (CADD) tools such as MOE, DataWarrior, and Pipeline Pilot exist.
For the automated workflow, the authors set out to develop a fragment library design tool with the following properties:
- Scalable, enabling the design of a large, diverse library with a small and focused set of screening compounds.
- An open-source computational workflow readily available for use by the scientific as well as the industrial communities.
- The ability to apply a wide variety of filters, such as costs, novelty, and physiochemical properties.
- To incorporate the 3D characteristics of the representative compounds.
The Workflow outline
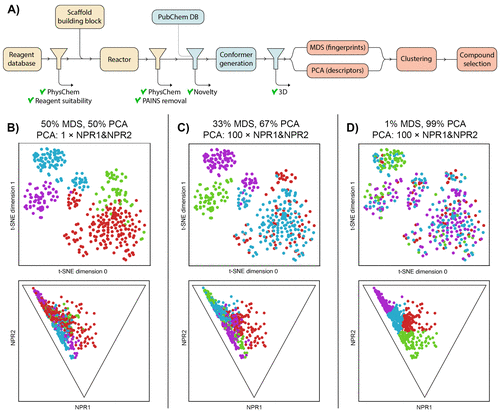
The authors used the KNIME ( Konstanz Information Miner ) open-source platform for workflow construction. KNIME enables the navigation of different properties and requirements necessary for the first-of-its-kind automated 3D fragment library design. KNIMe comprises nodes that are assigned specific tasks that can be linked in a sequence to generate a workflow resulting in the final output. The broad spectrum of available chemoinformatics nodes renders the platform a suitable choice for fragment library design workflow construction.
The workflow is divided into three stages:
- The generation of a virtual library involves importing reagent databases, followed by in silico reactions with predefined scaffolding blocks in a combinatorial manner.
- The second stage is novelty and shape analysis, which involves a comparison of the virtual generated library with the database of compounds encompassing the common scaffolds retrieved from the PubChem database using the PubChem Power User Gateway (PUG). The already reported compounds are then removed. The shape analysis involves using the RDkit conformer generator.
- The last and final stage is the selection of compounds from this virtually generated library, which involves clustering approaches incorporating maximum diversity.
Application of the workflow: A fragmented Library generated
In order to demonstrate the applicability of the workflow, the authors designed a set of fragments based on the 1,2 distributed cyclopropane scaffold. This resulted in a focused 10-member fragment library with a favorable physiochemical profile and values in the lower ranges within the RO3 limits for most of the calculated properties.
Conclusion
The authors have developed an automated workflow for the hassle-free construction of fragment libraries or fragment-based drug discovery using the KNIME platform. This workflow generates libraries with 3D screening compounds, whereas most fragment libraries found today are dominated by flat fragments. The incorporation of 3D structures results in novel drug candidates, and this approach will certainly lead to the development and design of several future drugs. Broadly, the workflow can be used for the construction of both small and large compound libraries.
Article Source: Reference Paper
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.