Scientists from the University of Saskatchewan, Canada, have performed a benchmarking analysis of the self-supervised contrastive learning methods used in image-based plant phenotyping. Plant phenotyping, traditionally performed manually, is now being handed over to deep learning-based methods. However, there is still a scarcity of methods for plant phenotyping tasks such as detection and counting. The authors try to fill this gap and benchmark two self-supervised learning (SSL)-based methods, viz., momentum contrast (MoCo v2) and dense contrastive learning (DenseCL), against the conventional supervised learning method for image-based plant phenotyping tasks. They find that supervised pretraining usually outperforms self-supervised pretraining and also show that MoCo v2 and DenseCL can learn different high-level representations as compared to the supervised method.
What is plant phenotyping?
Plant phenotyping is a crucial step involved in crop breeding. Plant phenotyping involves detection tasks as well as counting tasks, and the plant breeder is typically assigned the tasks of crossing parents and producing progenies with desirable and better traits. These desirable traits pertain to better yield and resistance to biotic and abiotic stress. Phenotyping accounts for a major bottleneck in the crop breeding process owing to the significant number of progenies involved and the chances of missing an improved progeny during the selection process. This is a highly labor-intensive task, and with the growing demand for accelerating the crop breeding process due to climate change and the ever-increasing human population, there is a need for automation in the plant phenotyping domain.
Image-based plant phenotyping
Image-based plant phenotyping has proven to alleviate the phenotyping bottleneck significantly. It involves capturing plant images over a growing season and using one or more imaging technologies. The images are then subjected to image analysis tools, and features or plant traits are extracted in a non-destructive and semi-automated way. This procedure is well implemented in the article by Rahaman et al. This method, as well as the platform developed by Das Choudhury et al., are both cost-effective and facilitate the measurement of plant phenotypes in a short time frame. However, they generate large amounts of high-throughput data, which makes complex feature extraction a challenging task.
Deep learning-based approaches for plant phenotyping: a plausible approach
The deep learning paradigm seems to be a plausible choice for devising automation in plant phenotyping. Deep learning techniques have been successfully implemented for image-based tasks such as classification tasks, object detection, as well as semantic segmentation. Several methods for plant classification, plant detection, plant stress classification, plant disease detection, and plant organ detection/counting have been developed using deep learning techniques. These are based on the supervised learning paradigm, which involves large datasets that are time-consuming and expensive.
The above limitation can be circumvented using the transfer learning paradigm, which is already a popular approach for image-based tasks. In plant phenotyping, transfer learning has already been implemented in methods for weed classification, plant detection, and plant center localization. The transfer learning workflow is not the ideal choice of approach for plant phenotyping, however. This is because such phenotyping tasks involve a source task trained on a large annotated dataset, which is not easily available for phenotyping.
Self-supervised learning (SSL) and contrastive SSL methods for plant phenotyping
The self-supervised learning paradigm combines aspects of both supervised and unsupervised learning techniques. They enable within-domain transfer learning and reduce the need for labeled data. Contrastive learning is an SSL paradigm that learns what makes two things different in a self-supervised manner. SSL-based contrastive learning approaches momentum contrast, MoCo v1/v2/v3 are quite successful in generating image-level features. Methods such as dense contrastive learning (DenseCL) have been developed for dense prediction tasks such as object detection and semantic segmentation.
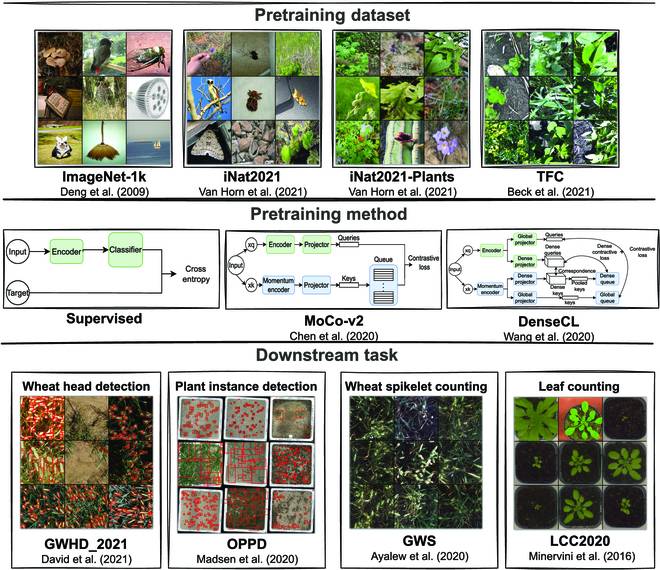
In the current article, the authors perform a benchmarking study that involves benchmarking the transferability of contrastive SSL methods against traditional supervised pretraining methods. The study involves the comparative analysis of four downstream image-based phenotyping tasks, viz., wheat head detection, plant instance detection, wheat spikelet counting, and leaf counting.
The comparative benchmarking analysis: a brief overview
The authors explore the benefits of using the DenseL approach over the SSL MoCo approach, MoCo v2, which learns global representations. The authors also investigate the role of the source domain in the transfer learning task using four different source domain datasets, viz, ImageNet, iNaturalist 2021, the Plants subset of the iNaturalist 20221 dataset, and the Terrabyte Field Crop (TFC) dataset.
With this backdrop, they performed a large set of SSL experiments and found the following:
- Supervised-pretraining results in the best-performing model on all the downstream tasks except the leaf counting task, as compared to MoCov2 and DenseCL.
- Domain-specific yet diverse pretraining datasets yield the best performance in downstream tasks across different pretraining methods.
- The SSL methods are more sensitive to redundancy in the pretraining dataset as compared to the supervised methods.
- There exists a high degree of similarity between the internal representations of models in MoCov2 and DenseCL across all layers. However, internal representations for supervised vs. self-supervised, although similar in the beginning, start to diverge towards the last few layers.
Conclusion
The authors set out to benchmark SSL contrastive learning methods for plant phenotyping for better development of future methods in this domain. They find the SSL paradigm-based approaches to be a promising approach for plant phenotyping tasks by leveraging unlabeled datasets. Although the authors caution against certain limitations as well. Lack of hyperparameter tuning may not yield optimal results with datasets other than ImageNet and could be addressed in future approaches with the rigorous tuning of hyperparameters. Nevertheless, the deep learning-based approach for plant phenotyping will certainly revolutionize the crop breeding process and provide solutions to several agricultural challenges posed by climate change and the growing world population.
Article Source: Reference Paper
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.