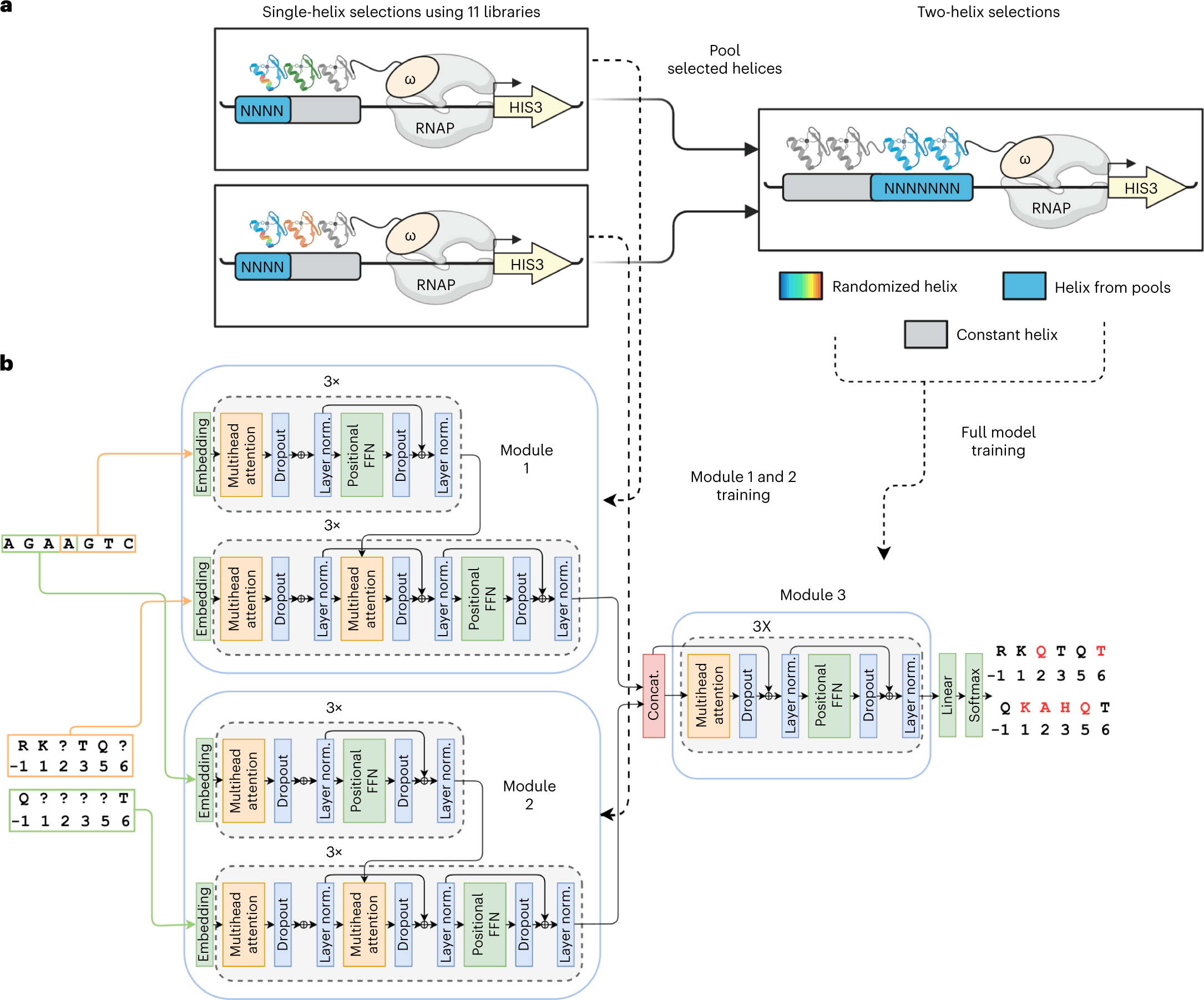
Controlling gene expression by engineering Cys2His2 zinc finger (ZF) domains to bind particular target sequences in the genome has many therapeutic applications. However, due to their intricate structure, designing these domains to interact with DNA is challenging. To address the problem, scientists from the University of Toronto and New York University have developed a deep learning based model, ZFDesign, that can solve zinc finger design for any genomic target. ZFDesign is an ML-based gene-editing method implemented using a novel hierarchical transformer architecture. The model is shown to perform better in terms of its universality in generating ZF designs.
Gene expression is controlled by various protein switches, like repressors and activators, via DNA binding in a sequence-specific manner. Among the diverse structural families of DNA-binding proteins, the Cys2His2 zinc finger motif is the most frequently used, starting from yeast to humans, and the mechanism of its binding to DNA is well understood.
The secondary structure of Zinc fingers comprises an α-helix and two adjacent β-sheets. The DNA binding results from α-helices being placed into the major groove facilitating specific amino acids to engage in base-specific contacts.
Given the widespread usage of the Cys2His2 ZF for DNA binding, engineering ZF domains to bind specific target sequences in the genome is an effective step toward programming gene expression regulation. This can lead to several potential therapeutic applications in curing diseases caused due to gene misexpression, haploinsufficiency, or gain-of-function mutations.
The various modes of engagement of ZF domains with DNA have made it challenging to design them.
Tools based on CRISPR-Cas and TALE (transcription activator-like effector) developed for such therapeutic applications are limited by the sheer size of the proteins that need to be delivered, among other intrinsic characteristics. Also, SpCas9 has been reported to have immunogenic risks upon long-term expression. On the other hand, ZFs require very few amino acids (<170) to specify a unique sequence in the human genome, which solves the delivery problem. ZFs have also been shown to be less immunogenic. Thus, ZFs prove to be a suitable candidate for programming the regulation of gene expression.
The Zinc Finger Design Model
The advent of language models in biology has resulted in remarkable technological advances. From generating novel protein sequences to novel protein structures, ML-based deep neural networks have paved the way for further novel discoveries. The recent addition to the bag of Deep Learning based models enabling the generation of novelty is ZFDesign.
The authors screened 49 billion protein-DNA interactions to generate codes and rules to design ZFs in a DL-based model ZFDesign. The MLbase model is designed for universal targets considering the compatibility of neighboring fingers and differences induced by a range of library environments. The model is implemented using a novel hierarchical transformer architecture.
Given the vast pool of screening data, an NLP-based approach seems fitting in capturing intricate details such as the neighbor finger influences. The model also uses the comprehensive single-finger library selections describing specificity amongst various finger neighbors and the 200 pair selections denoting ZF compatibility amongst neighbors. Given the hierarchical nature of the data, the authors developed a neural network with hierarchical attention modules.
- The model architecture is the traditional encoder-decoder. Encoders generate a high dimensional representation for each DNA base. Decoders predict each residue in a ZF helix.
- This is achieved using self-attention and attention layers relating nucleotide bases to helical residues.
- The model training includes the nucleotide target sequence as well as a partially masked ZF sequence, a masked language model.
The following figure illustrates the different layers in the neural network for the model for the specific case of Bacterial one hybrid(B1H) selection data.
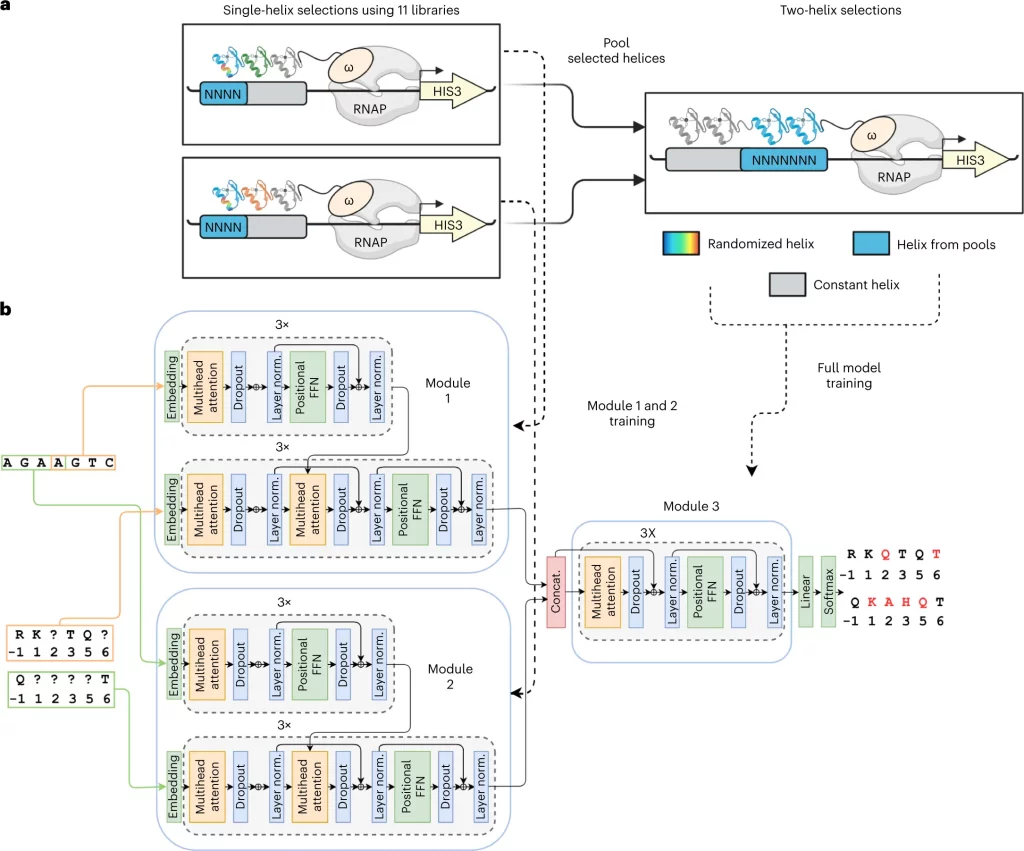
Image Source: https://www.nature.com/articles/s41587-022-01624-4/figures/3
Power of ZFDesign
A general strategy for engineering ZFs with novel specificity includes tweaking fingers one by one using functional variants from ZF libraries with randomized base-specifying positions of the helix. The other approach is based on the adjacent ZF interfaces resulting from neighboring finger influences. This results in innumerable combinations to be dealt with in building a design code for ZFs. ZFDesign implements a combined strategy that includes both, resulting in an exhaustive repertoire of general and interface-specific ZF designs. The authors find that while most targets show general binding strategies, specialized binding strategies exist for certain library selections. They also report that G(Guanine)-rich binding helices are found to be promiscuous. Comparison with the previously developed method ZFPred shows that the hierarchical model of ZFDesign performs significantly better. The authors also extensively report how human transcription factors could be seamlessly reprogrammed using ZFDesign, generating reprogrammed transcription factors (RTFs). Needless to say, this is a remarkable finding for therapeutics.
Conclusion
NLP-based language models have paved the way for technological advancements in producing artificially designed protein structures and sequences. ZFDesign is the new entrant in the series of such methods. ZFDesign proves to be a universal design method that produces novel ZF designs for general as well as specific gene targets. In comparison to previous methods, ZFDesign produces ZF array as nucleases, repressors, and activators, across a vast number of targets with high efficacy. The generation of RTFs is a remarkable feat in itself. The activation and repression activities of these designed RTFs are seen to be similar to those of CRISPR-based tools. The authors caution that the method, however, has limitations in terms of design, mostly due to G-rich binding promiscuity. This gives rise to off-target binding and hence unintended changes, which they hope to address in future iterations of the method. Given the vast repertoire of artificially designed ZFs at disposal, this will open up new avenues, both in research as well as therapeutics.
Article Source: Reference Paper
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.