Scientists from the University of Cambridge present Multifideity PubChem Bioassay, MF-PCBA, a collection of 60 molecular datasets with multiple modalities, reflecting the real-world nature of HTS (high-throughput screening) projects in drug discovery. MF-PCBA is a curated collection of datasets with two data modalities for each dataset, viz., primary and confirmatory screening, termed multi-fidelity. Previously, the same group presented 23 multi-fidelity HTS datasets from PubChem for benchmarking and included more than 6.1 million data points. The new collection comprises additional datasets and over 16.6 million protein-molecule interactions. Existing machine learning models for HTS data largely ignore noisy experimental measurements from primary screening. The authors envision multi-fidelity data and models as a natural step forward for early-stage drug discovery based on HTS data.
High-throughput Screening in Drug Discovery and the need for MF-PCBA
High-throughput screening (HTS) is one of the prime techniques in drug discovery. HTS identifies promising drug candidates in an automated and cost-effective manner. The set of largely automated techniques is applied to experimentally determine relevant biochemical interactions for a large collection of compounds. Hit identification in drug discovery through HTS is done in two phases, primary and confirmatory. The resulting data is multi-fidelity comprising noisy primary data for a large number of compounds and higher quality confirmatory data for a lesser number of compounds.
Existing computational pipelines do not take into account multi-modalities of HTS datasets and ignore noisy data from the primary screening phase. This results in millions of potentially useful data points being unused in models of bioactivity prediction. This enormous waste of such useful information prompted the authors to curate HTS datasets with multi-modalities.
Why do we need Machine learning benchmarks for drug discovery using HTS?
Machine learning approaches have revolutionized the curation of large datasets in the physical and life sciences. Deep learning techniques such as graph neural networks (GNNs) have emerged as the leading ML paradigm for learning directly on the data types found in the life sciences. The ability of GNNs to model complex relationships among objects has been vastly explored in quantum mechanics, particle physics, fluid dynamics as well as drug discovery. However, such approaches are dependent on high-quality datasets, and a lot of research efforts are currently directed toward developing high-quality datasets for graph representation learning.
Publicly available molecular benchmarks exist for several disciplines, such as MoleculeNet, Atom3D, and QM9. However, there is a lack of Machine learning benchmarks for drug discovery as well as HTS datasets with multi-fidelity. The development of such datasets would enhance the chances of innovations in early-stage drug discovery.
Precursor to MF-PCBA
The authors in a previous article have presented a compilation of 23 multi-fidelity HTS datasets from PubChem for benchmarking. This collection comprises more than 6.1 million data points, and the authors developed machine learning models for assessing the improvements offered by the integration of multi-fidelity data. The ML approaches included both classical machine learning as well as novel deep learning approaches based on GNNs taking into account both primary as well as confirmatory phase data.
Multifidelity-PubChem BioAssay (MF-PCBA)
With the background of the previous work done in the direction of compiling multi-fidelity HTS datasets for benchmarking, the authors set out to further expand the curated collection. MF-PCBA has 60 datasets with over 16.6 million data points representing unique protein-molecule interactions. MF-PCBA is by far one of the largest curated collections of graph-level representation datasets with the capability of adapting to classification paradigms. The usefulness of the multi-fidelity aspect of MF-PCBA is further validated by the authors using performing metrics such as R2 and mean absolute error (MAE).
The following steps are involved in the workflow of MF-PCBA development:
- The first step involves filtering the datasets with muli modalities such as molecular structures( represented by SMILES ), PubChhem CID for each molecule, SD( single dose), and DR (dose-response). This step involves using RDKit.
- The multi-fidelity integration workflow implements modeling confirmatory data (DR) using machine learning techniques such as Support Vector Machines (SVM) and GNNs.
- The primary screening data (SD) is learned separately and is incorporated through paradigms such as transfer learning by using graph or molecular embeddings.
- The embeddings are included in the DR step using augmentation.
The following figure illustrates the MF-PCBA workflow.
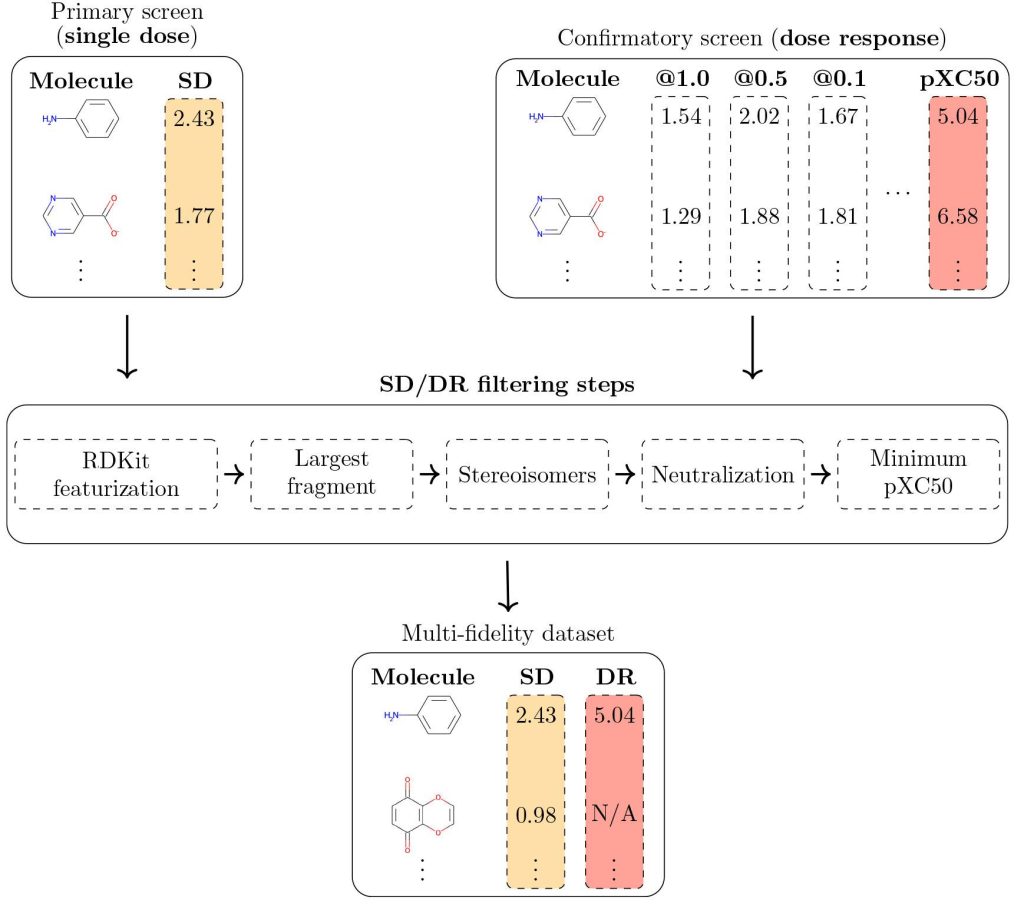
Image source: https://pubs.acs.org/doi/10.1021/acs.jcim.2c01569
Conclusion
The authors have developed a curated collection of 60 high-quality HTS datasets with multiple modalities that will aid in benchmarking machine-learning approaches for drug discovery. The multi-fidelity aspect has been shown to improve the ability to identify potential candidates for drug discovery. The inclusion of both primary and confirmatory phase data from HTS datasets will now enable the development of better computational pipelines for early-stage drug discovery. Such efforts as this are indeed remarkable in the field of big data and will aid the pharmaceutical community as well as the research community for the development of novel innovations in drug design as well as for the benchmarking of future methodologies.
Article Source: Reference Paper
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.










