Scientists from The Hospital for Sick Children, Toronto, have developed an atlas for childhood cancer diagnostic classification using machine learning approaches. Pediatric cancers are one of the leading causes of mortality in children worldwide. However, there is no comprehensive molecular assay for pediatric cancer diagnosis and classification. Currently available transcriptome-based diagnostic tools are based on supervised machine learning approaches that use pre-existing tumor labels. The authors developed RACCOON, a clustering approach for the unsupervised classification of cancer tumor subtypes, using RNA-seq data. Next, they developed a classifier for pediatric cancer, OTTER. Together, these methods generate an atlas for pediatric cancer classification.
Why do we need a novel methodology for pediatric cancer classification?
Childhood cancers differ from adult cancers as they develop mostly from embryonic tissue, thereby affecting several cell types. Childhood cancer prevalence according to subtypes also varies from adult cancers. Around one-third of pediatric cancers are leukemias. While carcinomas are common in adults, childhood cancers like neuroblastoma, a heterogeneous form of cancer, are rarely found in adults. Thus, pediatric cancer diagnosis requires a completely different and separate toolkit from the existing ones for adult cancers.
A comprehensive molecular assay for the diagnostic classification of pediatric cancer was the need of the hour. Transcriptome-based tools are a better choice than genome-sequencing-based tools owing to the fact that genome-sequencing data provides pointers to mutations preceding the tumor’s malignant transformation and does not account for current phenotypic variations. On the other hand, RNA-sequencing data is representative of the tumor’s current expression profiles, and methods based on RNA-seq can differentiate between tumors, regardless of their genomic origin.
Currently available transcriptome-based tools are based on supervised machine learning using pre-existing tumor labels and are not best suited for childhood cancer classification. In childhood cancers,intra-tumoral transcriptomic profiles tend to vary and can be largely pronounced, resulting in both poor as well as favorable prognostic signatures within the same tumor. Compared to adult cancers, childhood cancers have been found to have more transcriptional variation both within and between tumors. The authors exploit this transcriptional variation to develop a novel platform for pediatric cancer classification.
The authors developed a scale-adaptive clustering approach using RNA-seq data to classify tumor subtypes by implementing an unsupervised machine learning paradigm called RACCOON. The authors also developed a pediatric cancer classifier, OTTER. Together, they generate an atlas for childhood cancer tumor subtypes as well as a platform for the diagnostic classification of pediatric cancers.
RACCOON: Resolution-Adaptive Course-to-fine Clusters OptimizatiON
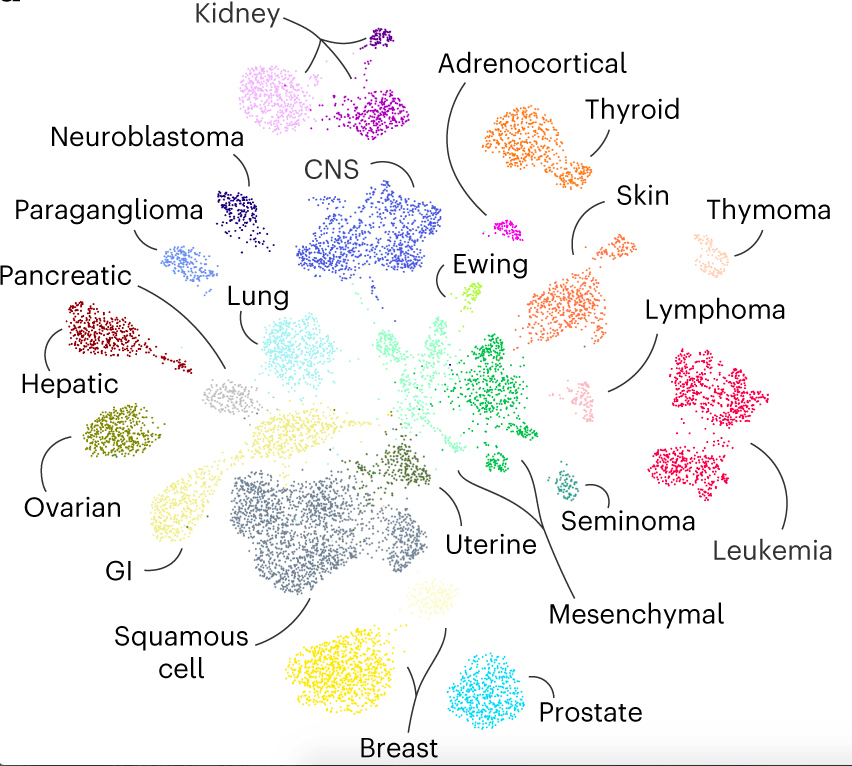
RACCOON is a novel, scale-adaptive clustering framework. The authors developed it to build an extensive reference hierarchy of tumor and normal subtypes. The methodology involves automatic optimization of parameters which aid in dimensionality reduction and low information filtering, which are essential for hierarchical clustering. The authors also ensure that the top-down building of hierarchies is not dependent on scale and dataset. RACCOON revealed 455 tumor subtype clusters representing 406 types of cancers from datasets comprising both childhood and adult tumor samples as well as non-neoplastic samples.
OTTER: Oncologic TranscripTome Expression Recognition
The hierarchical subtype clustering generated from RACCOON was next fed to OTTER, an ensemble of Convolutional Neural Network (CNN) classifiers. The algorithm reports probabilities of whether a tumor belongs to a class as well as its offspring classes, which generates a refined representation of a tumor’s subtype within a specific tumor lineage.
How good is it, really?
RACCOON identified 455 tumor subtype transcriptional classes representing 406 types of cancer. CNNs implemented in OTTER can match or refine pathologists’ diagnoses for 89% of patients. The method is not aware of the tumor site, morphology, or immunophenotype and is capable of classifying 90% of childhood cancers using RNA-seq data. The method performs robustly even when using a few million reads, a fraction of the RNA-seq data. The tool was able to find four subtypes of osteosarcoma with a clear difference in survival. The multiclass modeling approach results in revealing the expression of more than one subtype within a bulk tumor, such as was found in 50% of the neuroblastomas.
The following figure illustrates the atlas.
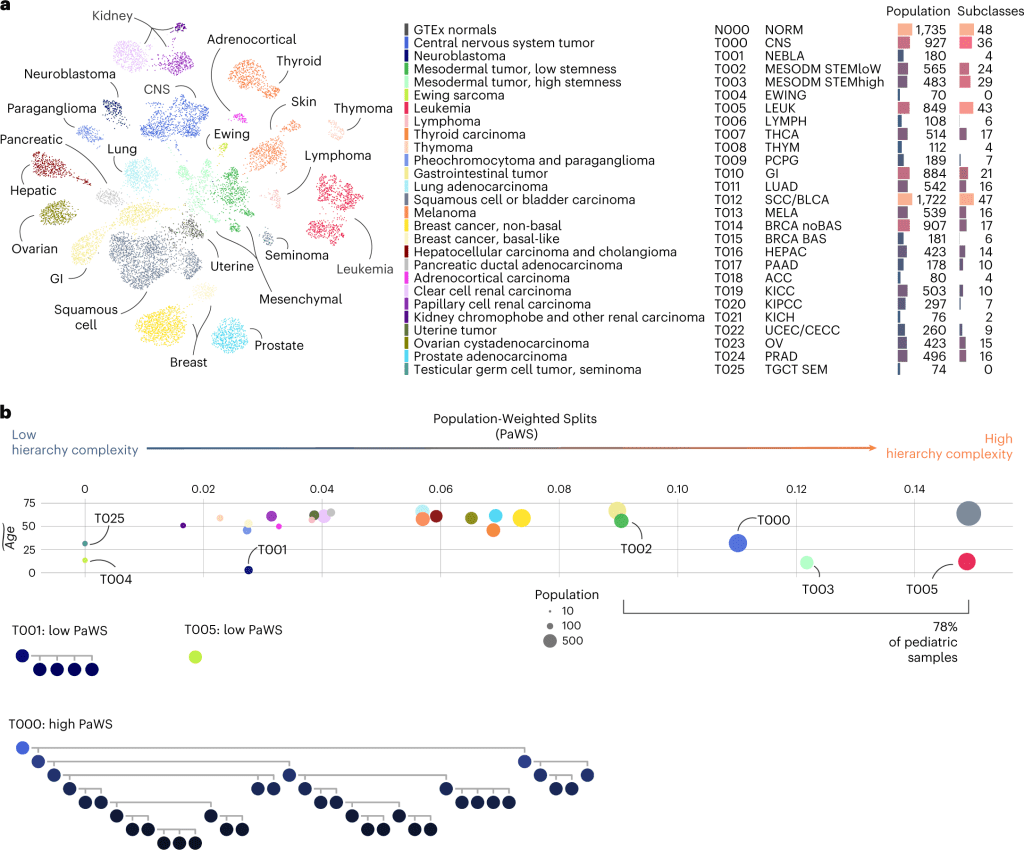
Image source: https://doi.org/10.1038/s41591-023-02221-x
Conclusion
Childhood cancer diagnosis and classification have met with several limitations in terms of tools and diagnostics, given the stark difference between childhood and adult cancers. Childhood cancer tumors show large transcriptional variations, and thus the need of the hour was to develop a transcriptome-based diagnostic tool for classifying pediatric cancers. The authors developed an atlas that comprises RACCOON and OTTER, two machine learning-based algorithms that classify childhood cancers with great accuracy. The authors quantify the unique transcriptional features of the cancer types and exploit this information to cluster and classify tumor subtypes and cancer types, respectively, in an unsupervised manner. The tool also has prognostic abilities, as was seen in finding osteosarcoma subtypes. The authors predict that with more data, this ever-learning tool will prove to be a diagnostic and prognostic tool for every child with cancer and aid in therapeutics based on tumor biology rather than histology alone. Needless to say, this atlas and the future iterations to come will aid the scientific as well as medical communities equally in the fight against pediatric cancers.
Article Source: Reference Paper | Reference Article | Transcriptional Cancer Atlas | Code Availability: RACCOON | OTTER
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.