Scientists from the Max Planck Institute for the Science of Light, Germany, have developed a novel methodology for rapid pathological analysis of cells from cancerous tissue samples using artificial intelligence and a single-cell approach. This methodology is capable of generating tumor biopsy analysis results in as early as 30 minutes, enabling intra-operative consultation pathology, which was previously time and resource intensive. The diagnostic methodology brings together enzyme-free mechanical dissociation of tissues, real-time deformability cytometry, artificial intelligence, and machine learning for data analysis. The authors also illustrate the usability of the method for the rapid examination of inflammatory bowel disease (IBD) samples.
The physical phenotype of cells and disease pathology
Physical properties of cells, such as cell size, shape, or deformability, are potential contenders for diagnostic and prognostic markers. Over the last decade, various tools have been developed, such as micropipette aspiration, atomic force microscopy and microbead rheometry, and optical traps for examining and analyzing the mechanical properties of cells. It is well established that a strong correlation exists between cell mechanical phenotype and the state of disease, as has been seen in the case of diabetes, sepsis, malaria, and cancer, among others. However, such methods are limited by low cell throughput as well as require specialist knowledge for usage.
Real-time fluorescence and deformability cytometry (RT-FDC): the game-changer
Real-time fluorescence deformability cytometry is a microfluidic technique that has high cell throughput and does not require specialist knowledge for usage. This is a label-free technique that has opened new doors for diagnostics. RT-FDC is capable of analyzing up to 1000 cells per second. In addition to being fast, the technique also provides multidimensional information, in addition to cell deformability, obtained directly from cell images. RT-FDC has proved to be a game-changer in diagnostics for diseases such as leukemia, bacterial infections as well as life-threatening Covid-19.
An intra-operative diagnosis of cancer biopsies
Malignancy characterization is largely achieved by solid tissue biopsy and is highly significant and useful for surgeons for intra-operative and peri-operative management of cancer patients. Histopathological analysis of frozen biopsy sections is the go-to methodology for solid tissue biopsy diagnosis. This conventional workflow involves several processing steps requiring pathological expertise. It is also time, resource, and labor-intensive. Alternative approaches, such as stimulated Raman spectroscopy, have been proposed but not yet implemented. Thus, an imminent need for a rapid diagnostic methodology for the pathological analysis of cancer biopsies.
Single-cell based AI pathologist
The authors have developed a rapid method for the processing and analysis of cells from solid tissue aiding biopsy-based diagnostics. The method incorporates RT-FDC along with artificial intelligence and machine learning techniques for extracting various physical phenotypic features of the imaged cells for deformability analysis. This is a label-free method that does not involve any expensive reagents and markers as compared to conventional cytometry workflows. The greatest advantage lies in how fast and rapid the methodology is. The information is available within 30 mins of biopsy excision and also does not require sample freezing or transportation of frozen samples. This is remarkable for detecting pathology quickly and saving patients from returning for surgeries, and even preventing death.
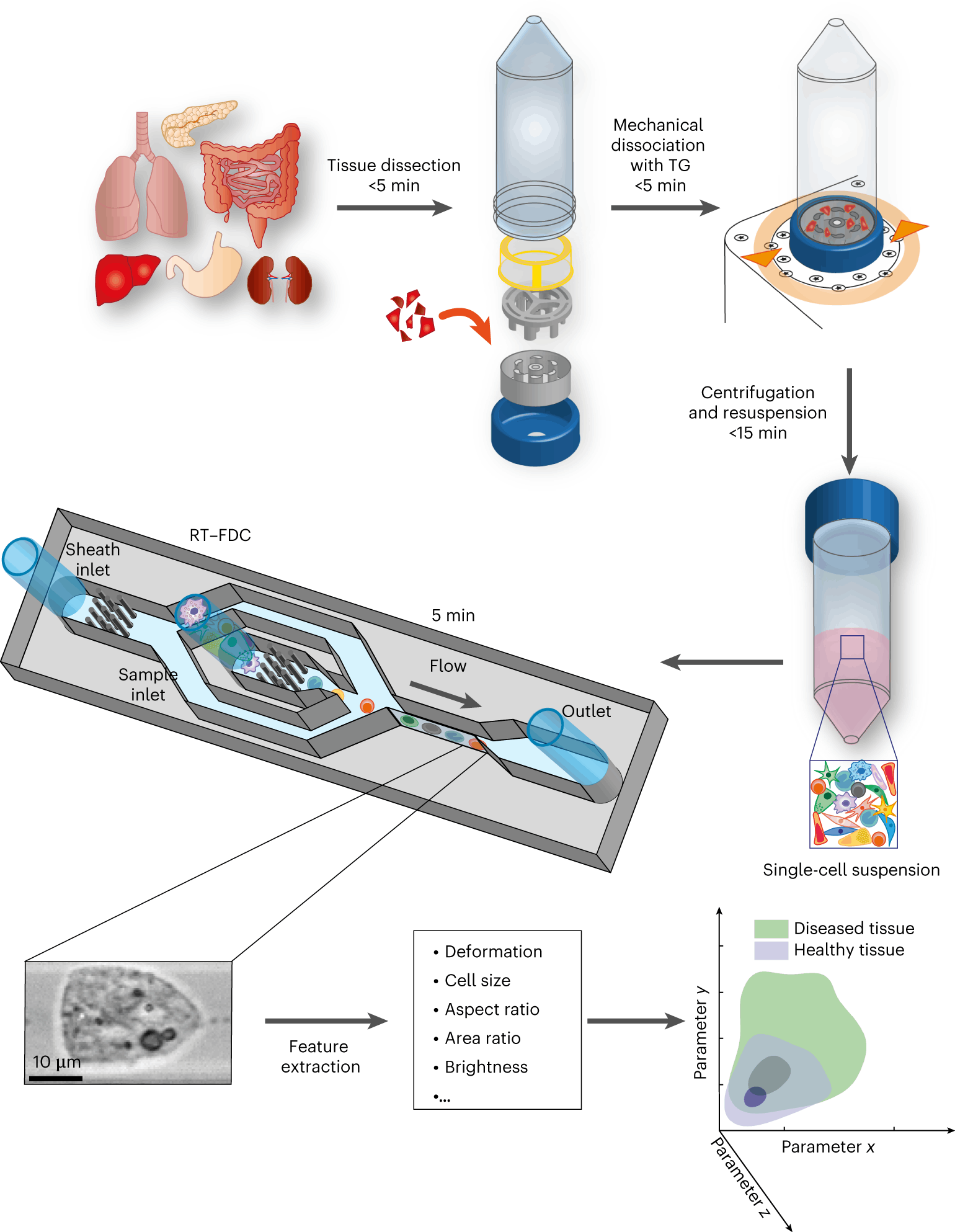
Physical phenotyping of cells from mechanically dissociated tissues involves the following steps:
- Dissection of the tissue sample is followed by mechanical dissociation.
- The next step involves the RT-FDC workflow.
- The final step involves the AI module that quickly analyses the large datasets generated by the RT-FDC workflow and performs a rapid assessment of whether a biopsy sample contains tumor tissue or not.
The following figure illustrates the workflow:
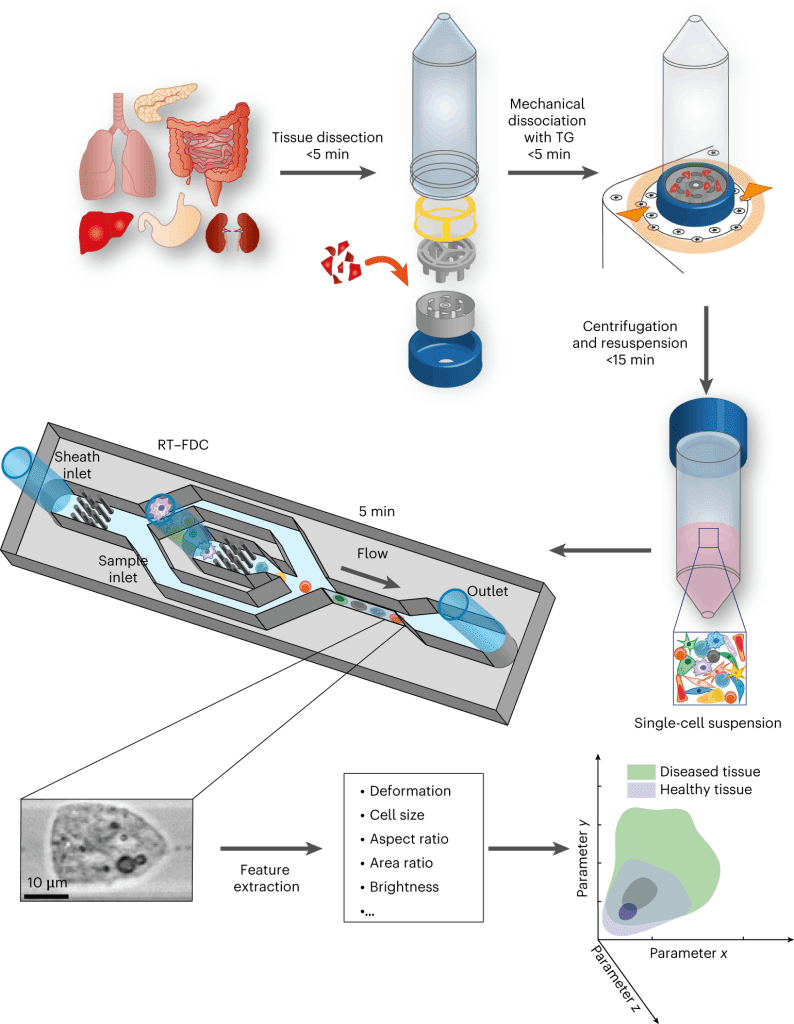
Image source: https://doi.org/10.1038/s41551-023-01015-3
The methodology has been applied to several diagnostic scenarios, such as the following:
- The cell physical phenotyping workflow is able to detect tissue inflammation, as in the case of Irritable Bowel Disease (IBD). The authors find that physical phenotypes of cells change upon inflammation, and it is well established that chronic and prolonged inflammation leads to malignancy.
- The authors next tested their method on cancer cells. It is well known that cancer cells differ physically from their healthy counterparts. The results revealed that the physical phenotype of cancer cells is remarkably different from healthy cells. They were found to have larger cell sizes and higher order of deformity as compared to healthy cells.
Conclusion
The development of a rapid diagnostic method for biopsy analysis has been the need of the hour for cancer treatment and therapy. However, histological and pathological procedures involved in the pathological diagnostic of tumor biopsy samples are time and resource consuming, thereby often not rendering early assessments as required. The authors developed a quick methodology incorporating RT-FDC and an AI-based approach that enables tumor detection from tissue samples as early as 30 minutes. The method has also been shown to detect inflammation in IBD owing to changes in the physical phenotypes of the diseased cells. This no longer requires a trained specialist to examine and assess the tissue samples for malignancy. In the future, the application of this methodology on larger patient cohorts will enable the design of a remarkable and groundbreaking AI pathologist.
Article Source: Reference Paper | Reference Article
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.