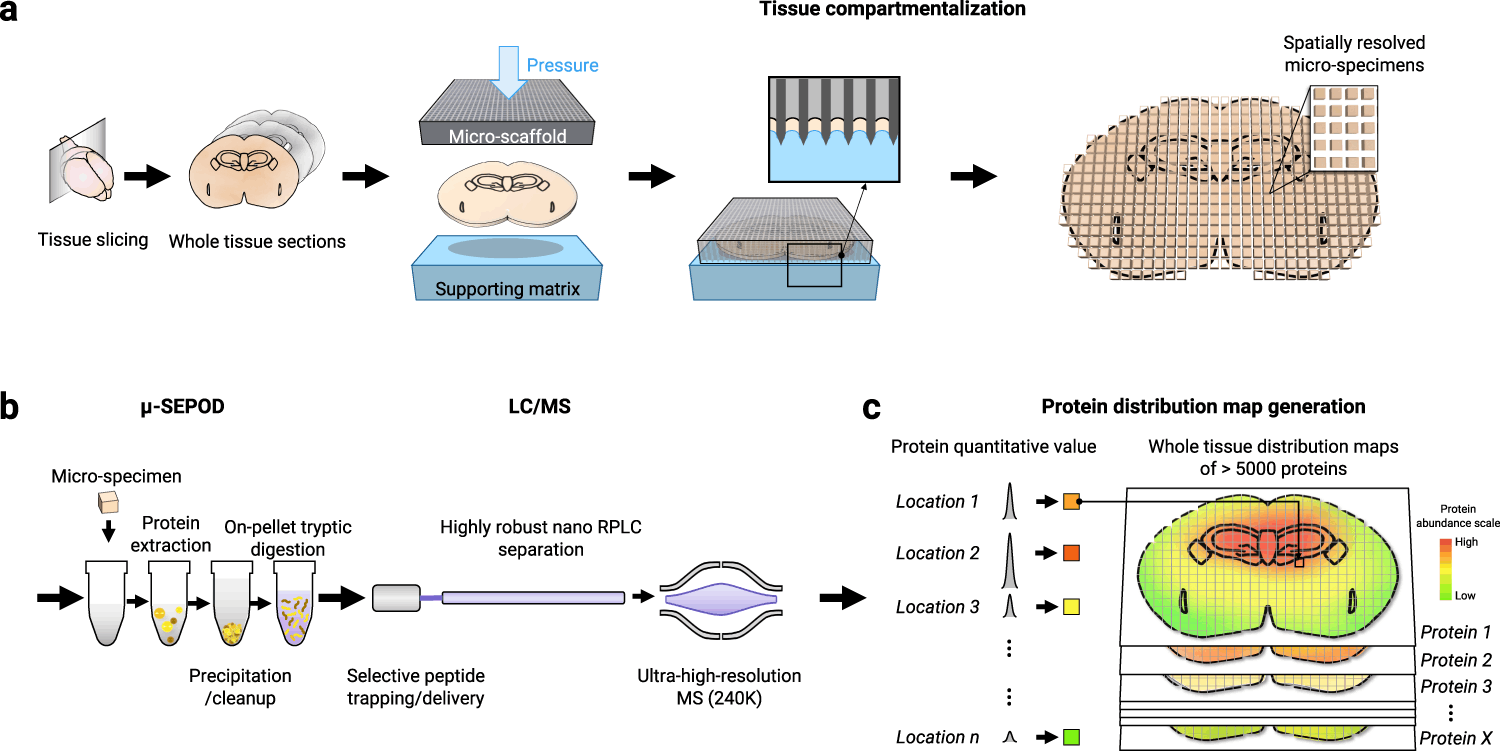
Researchers from the Jun Qu lab at SUNY Buffalo, New York, have developed a revolutionary method for in-depth mapping of protein localization in whole tissue. The method implements the cutting-edge technique of micro-scaffold-assisted spatial proteomics (MASP). The daunting task of accurate, in-depth mapping of protein localizations at the whole-tissue level has been achieved by using spatially-resolved micro-compartmentalization of tissue using a 3D-printed micro-scaffold. The MASP pipeline accurately maps thousands of proteins across a whole-tissue slice with high efficacy and precision. The MASP pipeline was successfully applied to map cerebral proteins in the mouse brain. This resulted in mapping a great number of proteins (>5000), some for the first time, including several important brain markers, regulators, and transporters.
Tissues are composed of specific cell types, which result in homogeneity across the tissue of interest. However, the protein distribution across this tissue is heterogeneous, and this compartmentalized and localized protein distribution forms a crucial part of several physiological processes and biological mechanisms. Quantitative characterization of these heterogeneous protein distributions enables a critical understanding of spatially regulated processes and networks spanning the entire tissue, which is essential for an informed understanding of disease mechanisms and therapeutic effects.
Spatial proteomics and why we need MASP
The proteome is the complete repertoire of proteins in a cell type or tissue, as the case may be. The advent of advanced mass spectrometric methods has enabled the determination of proteomes of numerous cell types and tissues. In today’s time, it is possible to identify and accurately and precisely quantify numerous proteins from tiny (nanogram) quantities of input material along with their post-translational modifications. However, this alone is not enough, as the spatial organization of proteins in cells and tissues is of great biological significance. The sub-cellular as well as tissue-level compartmentalization or localization of proteins is involved in various cellular and tissue-level mechanisms ranging from subcellular transportation to signaling and is the focus of Spatial Proteomics. Difficulties in the sensitive and quantitative mapping of the numerous tissue proteins pose a staggering challenge in obtaining spatially-resolved accurate proteome mapping. State-of-the-art methods, like immunoassay-based methods like multiplex-IHC and Matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI), all pose their own set of difficulties and challenges.
Liquid chromatography-mass spectrometry (LC-MS) based technologies and AI-based laser-microdissection (LMD)-assisted NanoPOTS imaging methods are promising candidates for proteome analysis. However, these methods are best suited for microscopic, focused tissue areas but are not suitable for generating whole-tissue level protein mapping.
The need of the hour is a method that facilitates whole-tissue proteome mapping with deeper proteome coverage than prevailing methods. The authors developed a novel approach, MASP, to address these shortcomings.
MASP was developed to end the search for a reliable and robust micro-sampling method, enabling uniform compartmentalization of whole-tissue slices.
The MASP pipeline
The pipeline comprises three components, each addressing the aforementioned shortcomings.
- Tissue micro-compartmentalization, which uses a 3D-printed micro-scaffold, generates precise compartmentalization.
- LC-MS analysis preceded by extraction, clean-up, and digestion of location-specific micro-specimens.
- Using the MAsP app, generate protein distribution maps.
Applications of the MASP strategy
The MASP pipeline was applied to healthy mouse brain cerebral tissue to obtain whole-tissue protome distribution. By applying stringent cut-off thresholds, 5019 proteins were identified and quantified. The pipeline generated complete distribution maps for more than 905 of the proteins, and these were found to be located in at least 95% of all locations. Validation analysis of the MASP-generated mapping proved to be quantitatively accurate and reliable.
MASP generates landscapes of spatially-organized signaling pathways and biological functions
MASP generates whole-tissue protein distribution maps for numerous proteins involved in signaling pathways and networks. This results in a comprehensive understanding of various spatially regulated and organized biological mechanisms involved in diseases and otherwise.
The following figure illustrates the protein distribution mapping for the KEGG pathways involved in the Dopaminargeic Synapse pathway.
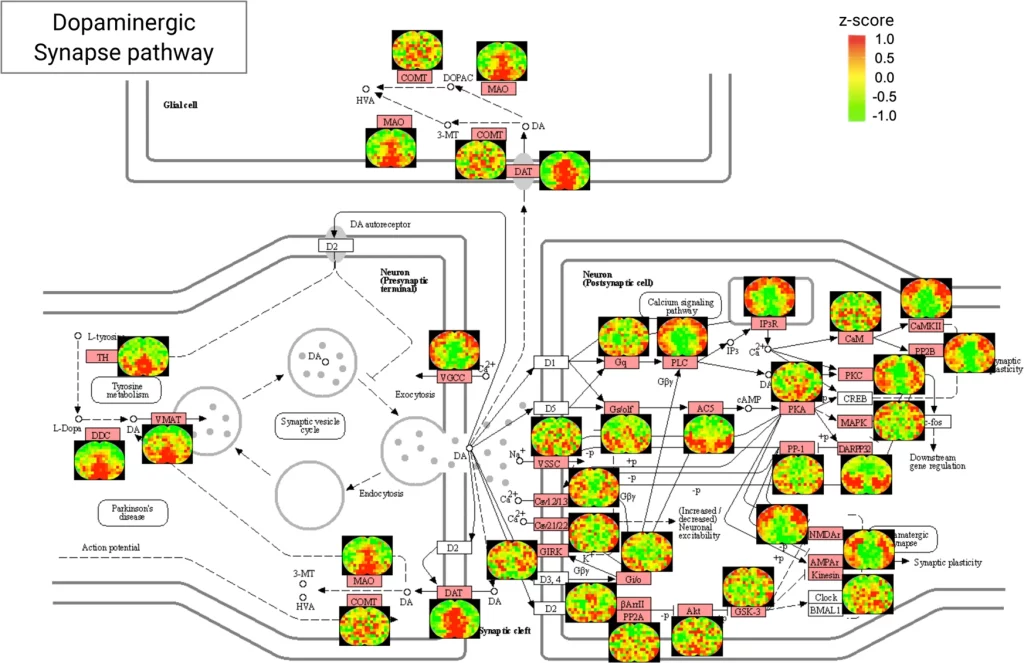
Image source: https://www.nature.com/articles/s41467-022-35367-2/figures/5
By means of several examples of different pathways involved in different brain regions, the authors generate maps for protein distributions in important synaptic pathways. These pathways are involved in various neurodegenerative diseases like Alzheimer’s disease and Parkinson’s disease, as well as aging and cognitive functioning. Needless to say, the generation of such reliable and in-depth protein distribution maps for these pathways will aid in the development of therapeutics for the said diseases. The authors also generate maps for pathways involved in neurotransmitter transport. Such an in-depth distribution of localized protein mappings for these pathways will aid in the phenomenological modeling of transport mechanisms in neurons.
Conclusion
In-depth, spatially-resolved mapping of the whole-tissue proteome is finally achievable with MASP. The MASP pipeline capabilities complement existing single-cell proteomics or LMD-based technologies in generating reliable and accurate whole-tissue protein distribution mapping. The 3D-printed micro-scaffold aided micro-compartmentalization strategy, the LC-MS analysis, and the MAsP app all do away with the challenges previously posed in unison.
The dataset generated by applying MASP to the brain tissue of healthy mice produced >5000 protein distributions, providing researchers with a vast repertoire of proteins in significant pathways, aiding in scientific research as well as drug development. Future developmental efforts in terms of higher spatial resolution are ongoing, which could lead to applying MASP for other molecules of interest.
Article Source: Reference Paper | Reference Spatial proteomics | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.