Scientists from the University of California, Irvine, have developed NeuronChat for inferring inter-neuronal communication using single-cell transcriptomics data. Previous cell-cell communication inference methods are based on short-range signaling enabled by diffusion or physical contact. Neurons communicate via synapses involving neurotransmitter signaling and extend over long distances. The connectome, the wiring organization of neural connectivity, is influenced by transcriptional regulation. Thus, understanding inter-neuronal communication requires spatial as well as transcriptional regulatory information. The authors designed NeuronChat, a method and package for the inference, visualization, and analysis of inter-neuronal communications from single-cell expression data as well as spatially resolved transcriptomics.
Cell-cell communication inference and why we need NeuronChat
Brain functions rely on signal transmission among neurons as well as non-neuronal cells. This signal transmission takes place over long distances, given the neuronal morphology. The neurons interact with each other at synapses via neurotransmitter signaling, which is largely influenced by the transcriptional regulatory machinery of the cell. Neurotransmitters, the chemical messenger of neurons, are often transported from the cell body of the neurons to the presynaptic terminal of the neuron upon neuronal activation. This requires concerted and coordinated action between several genes involved in these processes. Thus, it is necessary to include neurotransmitter signaling players as well as their actions while inferring inter-neuronal communication.
The heterogeneity of cell transcriptomic states, as revealed by the recent single-cell RNAseq (scRNAseq) datasets, allude to the possibility of different gene expression profiles across regions, implying different neural signal processing states. Spatial transcriptomic methods applied to neuronal datasets have generated cell atlases that measure the gene expression levels in cells coupled with the spatial location of neural cells. These transcriptome-level data naturally contain the gene expression levels for genes involved in neuronal signal transmission. Thus, incorporating scRNAseq data coupled with spatial transcriptomics data could be the determining factor for inferring inter-neuronal communication.
Previously developed cell-cell communication methods like CellChat, CellPhoneDB, and NicheNet are based on short-range autocrine/paracrine signaling enabled by ligand diffusion or physical contact between cells. While these methods are capable of determining cell-cell communication networks, they are not suitable for neuron-neuron communication inference for the above-mentioned reasons. Hence, the authors developed NeuronChat for the inference of neuron-neuron communication aided by single-cell transcriptomics information.
Overview of NeuronChat
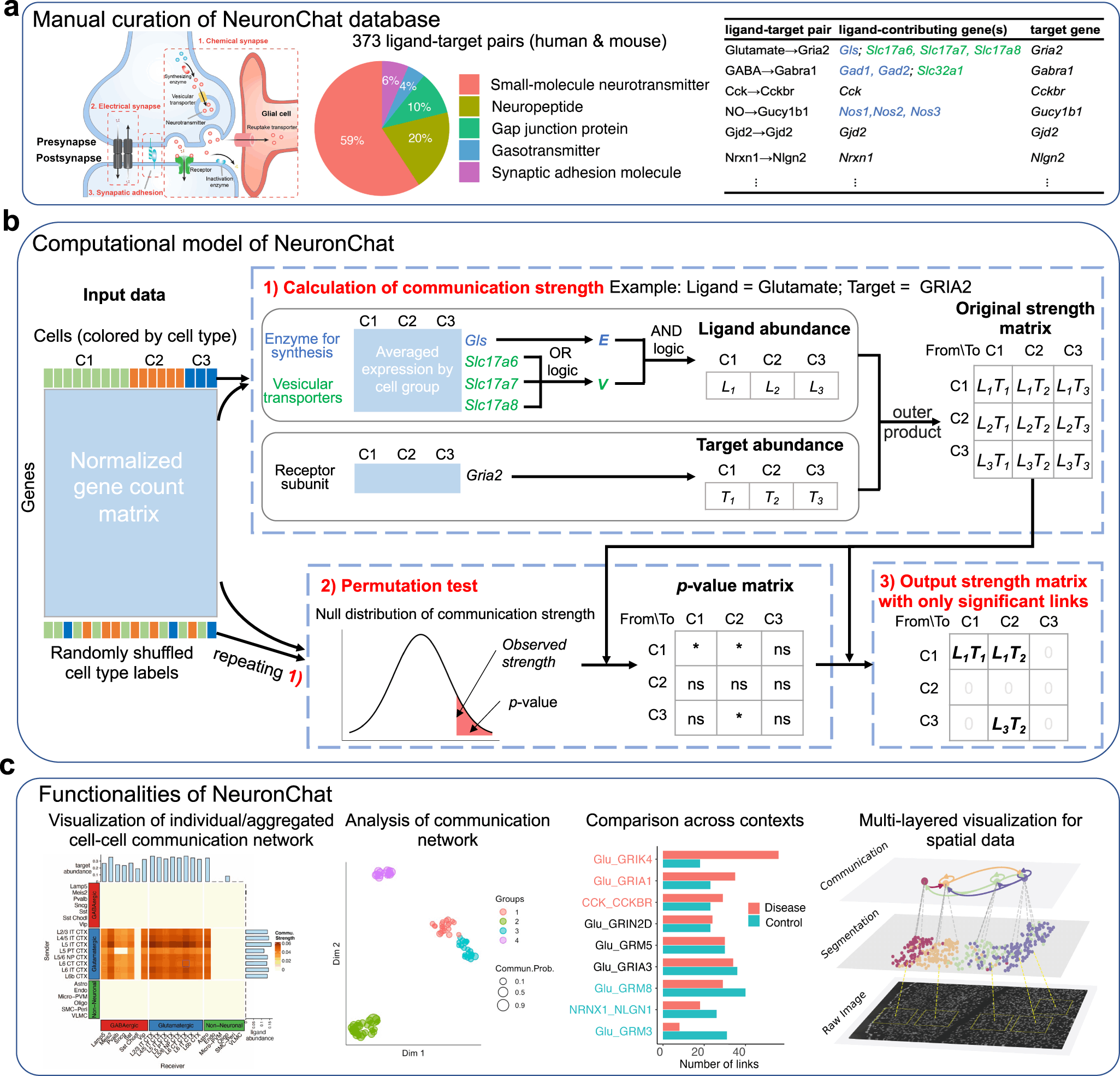
The authors first curated a neural-specific database NeuronChatDB containing intercellular molecular interactions for both mouse and humans. Each interaction pair contains a ligand and a cognate target, as well as genes involved in their synthesis and vesicular transport. For heteromeric receptors containing multiple subunits, individual subunits are curated separately with the same ligand.
The next step involves the construction of a computational model for linking the expression data to cell-cell communication probability. This involves the following three components:
- Calculating the ensemble average expression.
- Calculating the cell-cell communication strength.
- Determining the importance of communication links.
The following figure illustrates the above steps.
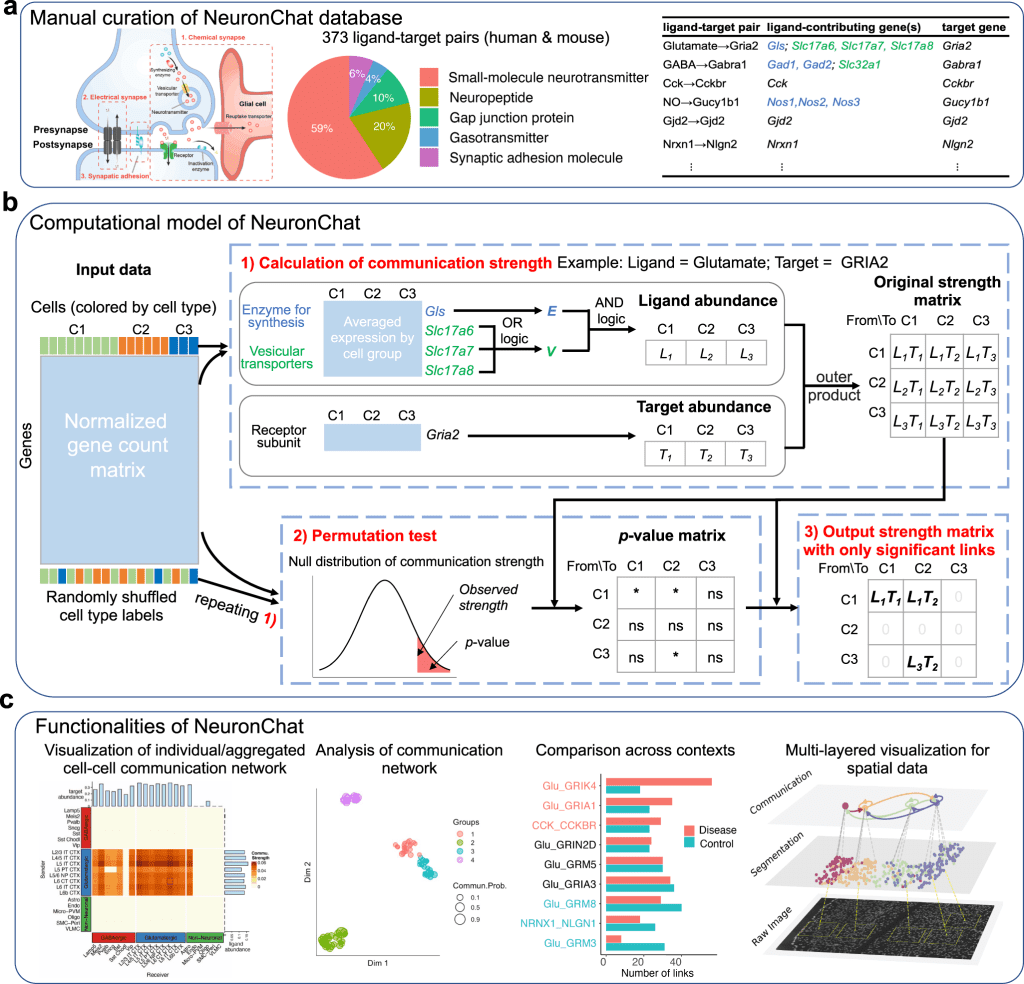
Image source: https://doi.org/10.1038/s41467-023-36800-w
Comparison with other cell-cell communication inference tools
The authors implement the same workflow for NeuronChat as in CellChat as well as CellPhoneDB. However, NeuronChat differs from the previous methods in terms of the calculation of the ligand abundance and the formula for calculating the communication strength.
NeuronChat models the ligand abundance by applying AND logic, which is the geometric mean among different groups of genes, while applying OR logic (arithmetic mean) for redundant genes within the same group.
Where as, in the other methods, small-molecule neurotransmitters are implemented without categorizing the gene into functional groups, thereby using only AND logic (geometric mean) for all contributing genes and no OR logic for the redundant genes.
NeuronChat identifies intercellular communication patterns
The authors applied NeuronChat to the scRNAseq data of the mouse primary visual cortex (VISp). NeuronChat identifies intercellular communication patterns as well as function-related interactions, as illustrated in the following figure.
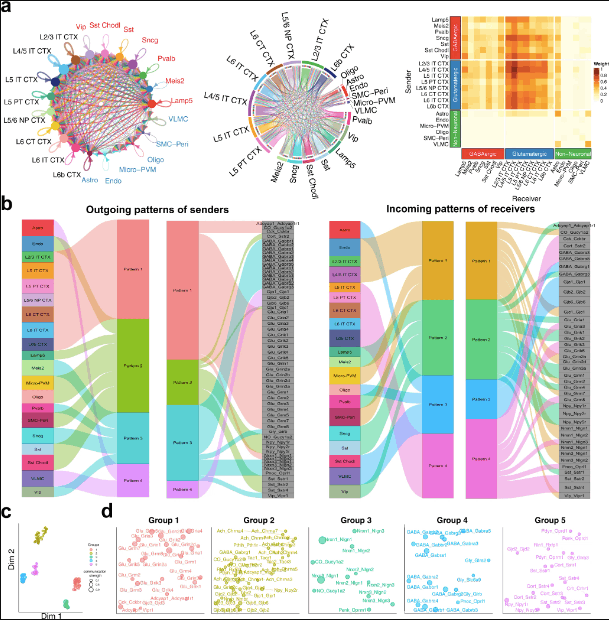
Image source: https://doi.org/10.1038/s41467-023-36800-w
NeuronChatDB has ligand-target information for humans. The author next applied NeuronChat to single-nucleus RNAseq data from cortical tissue from patients with Autism Spectral Disorder (ASD) and healthy controls. NeuronChat was able to predict the change of intercellular communication patterns in the patient cohort.
The authors apply NeuronChat to reveal conserved and context-specific communication patterns between interlaminar excitatory networks for ALM and VISp. They compare individual communications networks for both ALM and VISp using functional similarity. This is implemented using manifold learning techniques. This elucidates the capability of NeuronChat to categorize the detected ligand-target interaction pairs into biologically meaningful groups.
Conclusion
NeuronChat proves to be a strong contender for predicting neural connectivity using scRNAseq and spatial transcriptomics data. Novel neural connections among diverse transcriptomic states can be predicted using NeuronChat. By contrasting and complementing the NeuronChat connectivity predictions for particular ligand-target pairs with existing data from electrophysiology as well as retrograde labeling, one can identify key players in terms of genes and signaling pathways involved in uncharacterized mechanisms underlying neural connectivity. The NeuronChat R package provides versatile and user-friendly visualization tools as well as tools for network analysis. The NeuronChat model settings can be modified to further incorporate stoichiometric effects of metabolism and pathways dependency for the molecules in addition to the existing parameters. The authors also envision applying NeuronChat to proteomics and metabolomics for ligand-target interaction inference if the data is available. With its vast application scope and ability to predict inter-neuronal communications, NeuronChat will be a game-changer in fundamental research as well as for innovations in therapeutics.
Article Source: Reference Paper
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.