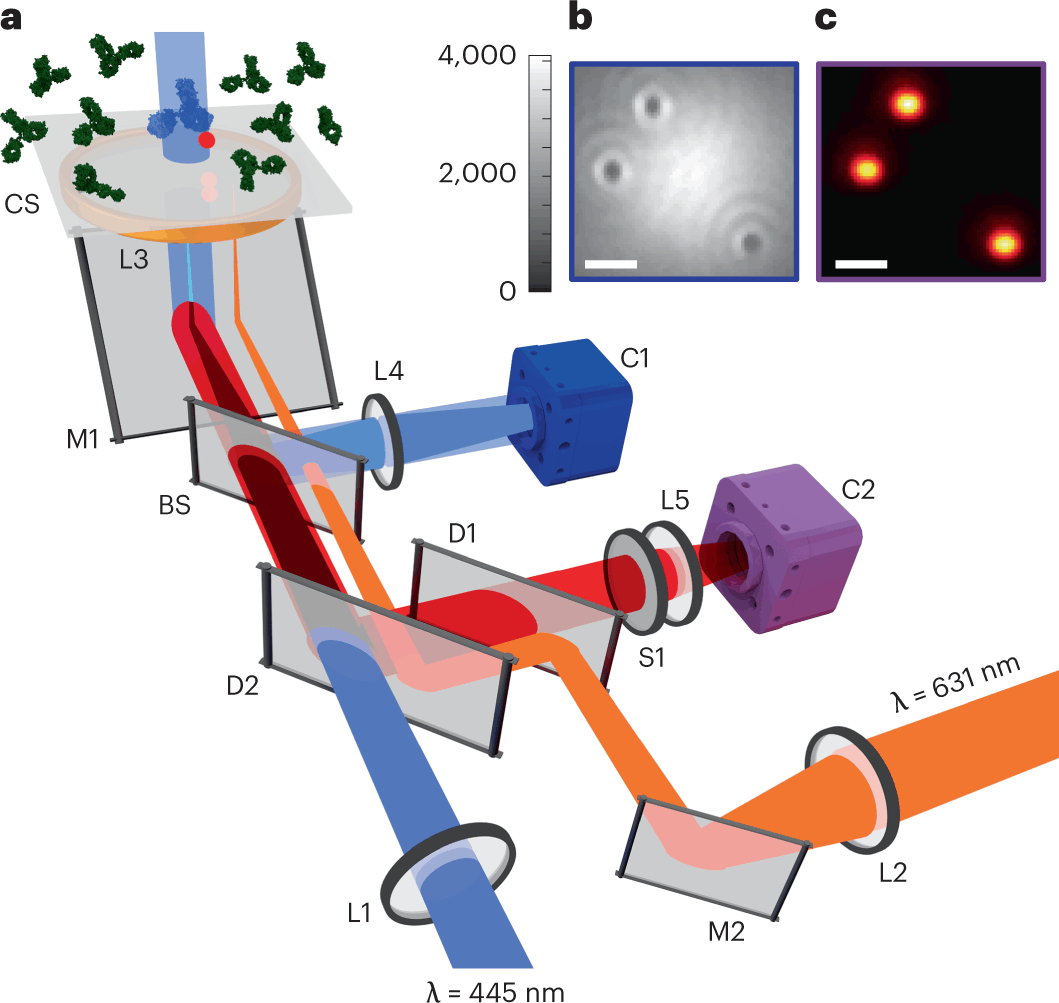
The detection of biomolecules at the nanoscale is of great significance in fundamental biology research, just as it is for biomedical investigations. The evolution of techniques for the detection and characterization of biomolecules has resulted in remarkable scale and resolution in terms of the size and mass of the molecules. Scientists from Germany have achieved the remarkable feat of pushing the sensitivity limits of interferometric scattering (iSCAT) microscopy, a label-free optical technique for the detection of proteins. The authors accomplish this using an unsupervised machine-learning algorithm and are able to detect proteins with a mass as low as 10 kDa, which is four times smaller than proteins being detected using earlier techniques.
Why do we need to detect tiny proteins, and how to go about it?
The detection and characterization of nanoscale matter are of utmost importance in the understanding of fundamental biological mechanisms involved in physiological processes as well as in diseases. The quest for the analysis of the matter at such scales and resolutions has led scientists and researchers to apply varied techniques to resolve molecular properties like structure, chemical composition, weight, size, and even molecular dynamics. Nuclear magnetic resonance spectrometry, electrophoresis, mass spectrometry, electron microscopy, fluorescence imaging as well as plasmon resonance spectrometry have yielded a plethora of information regarding biomolecules. However, each method has its own share of measurement limitations.
Optical methods seem to be the best fit in this regard for being non-invasive and compatible with real-time studies. Interferometric scattering (iSCAT) microscopy enables the detection of single proteins in a label-free manner. This method is capable of measuring protein mass as well as localizing protein binding positions with nanometer precision. Ideally, the technique is only limited by shot noise, which is the fluctuation in the number of photons detected. This implies that the sensitivity of iSCAT should be increased with more incident photons, and it should be able to detect molecules with arbitrarily low mass. But in practice, several technical noise sources coupled with speckle-like background fluctuations have limited the detection sensitivity of iSCAT.
Thus, the authors set out to develop a machine learning-based approach for improving the sensitivity of this technique and successfully implemented a machine learning algorithm to push the sensitivity limits.
Pushing the limit using machine learning
Machine learning techniques have already been in use in the field of microscopy for background correction or signal enhancement. Background correction typically implements techniques from computer vision using spatial and temporal information. Supervised Deep Neural Networks (DNNs) have been used for the extraction of spatiotemporal features in localization microscopy and particle tracking. Applying supervised techniques is limited by the lack of complete knowledge about the signal and noise properties. An attempt to get around this limitation by Sheth et al. used an unsupervised DNN based on FastDVDnet to denoise an image series.
The authors implement a tailor-made scheme for the self-supervised FastDVDnet. They first denoise their images and then obtain the PSFs (point spread functions) of the rare landing proteins by subtracting the denoised frame from the frame of interest.
The next step involves classifying the outcomes for which they use iForest, an unsupervised algorithm used in anomaly detection. Anomaly detection algorithms involve a common paradigm of first defining a normal signal and then identifying the deviations or anomalies. In this case, the normal signal is the residual background speckle image which is obtained by averaging over many frames that occur immediately before and after the frame containing a protein landing event.
The authors have also explored a tailor-made approach for gaining better insight into the underlying physical criteria in anomaly detection by choosing a set of temporal and spatial features that are calculated for a certain pixel range in an image frame. A feature matrix results from these one-dimensional frame values and is fed to iForest for classification.
The authors benchmarked and validated the results by performing fluorescence detection in total internal reflection (TIRF) mode.
With this methodology, the authors were able to detect tiny proteins with molecular mass as low as 21kDa, 18kDa, and 9kDa. The previously achieved scale in terms of size was 44 kDa. This is indeed a remarkable achievement in terms of detecting very small proteins.
The following figure illustrates the image frames with the detection of very small proteins.
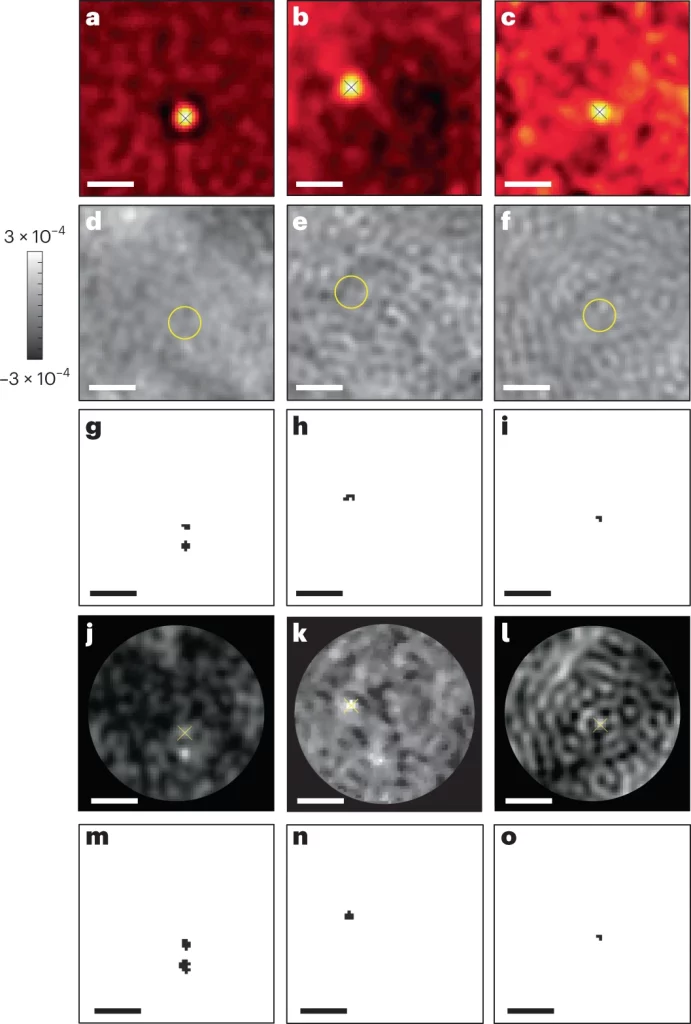
Image source: https://doi.org/10.1038/s41592-023-01778-2
Conclusion
The authors used a novel approach to detect very small proteins by applying a self-supervised machine-learning algorithm, which is used for anomaly detection, to iSCAT image frames. The resolution achieved is groundbreaking, as the authors were able to detect very small proteins with a molecular mass as low as 9 kDa. This is a huge improvement, as previous methodologies were able to detect proteins with a molecular mass as low as 40 kDa. Being a label-free, optical detection technique-based method, it enables real-time detection of these very small proteins. The biological significance of this improvement is huge, as the detection of such small molecules will now aid in our understanding of several physiological processes, such as the localization of proteins, protein trafficking, and many more. This method will also facilitate the understanding of abnormal protein aggregation in several life-threatening diseases. Needless to say, this method will find huge applications in drug designing and therapeutics as well.
Article Source: Reference Paper
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.