Scientists from the University of Toronto, Canada, have developed a foundation model for single-cell analysis, scGPT, by generative pre-training on over ten million cells. The generative pre-trained transformer for single-cell analysis is capable of capturing meaningful and crucial biological insights at the gene and cellular levels. The foundation model can be finetuned for numerous downstream tasks such as muti-batch integration, multi-omic integration, cell type annotation, genetic perturbation prediction, and gene network inference and has been shown to achieve state-of-the-art performance in these tasks. In the future, the authors aim to develop a pre-trained model capable of understanding different tasks and contexts in the zero-shot setting that does not require finetuning.
How can generative pre-trained models aid in single-cell analysis?
Generative pre-trained models such as DALLE-2 and GPT-4 have revolutionized the fields of computer vision and natural language generation (NLG). These foundation models are based on the paradigm of pre-training transformers on diverse large-scale datasets. Such models can be finetuned for numerous downstream tasks, and interestingly, they have demonstrated improved performance than task-specific models, which are trained from scratch.
Machine learning-based approaches in the domain of single-cell analysis have been restricted to model development dedicated to specific analysis tasks. The breadth and scale of the datasets involved in such approaches are also often limited owing to sequencing capacity as well as the scope of the research question. Thus, the need of the hour is the development of a foundation model pre-trained on large datasets that further our understanding of single-cell biology. In this regard, scGPT bridges the gap.
Generative pre-training in single-cell biology has not been vastly explored hence, the authors derive inspiration for applying this paradigm from other domains. Self-attention transformers have been shown to efficiently model input tokens of words. The authors use the analogy of words in texts to genes in cells in the sense that genes characterize cells the way texts are made of words. The authors envisioned the possibility of learning and extracting gene and cell representations simultaneously, as is possible in the case of word and sentence embeddings in NLG.
scGPT: Single-cell Generative Pre-trained Transformer
The foundation model for single-cell analysis, scGPT, has stacked transformer layers at the core of the model. These stacked layers have multi-head attention implemented that learns cell and gene embeddings simultaneously. The workflow comprises two stages: the pre-training stage and the finetuning stage. The pre-training stage incorporates 10.3 million scRNA-seq data from blood and bone marrow cells obtained from the CellXGene portal for training. sGPT learns in a self-supervised manner using a specially designed attention mask and generative training pipeline, which enable the model to gradually learn and generate gene expression in cells based on simple cell or gene cues.
The finetuning stage is where the users can apply the pre-trained model to new datasets and specific tasks. The authors developed pipelines for downstream tasks such as scRNA-seq integration with batch correction, cell type annotation, multi-omic integration, perturbation prediction, and gene regulatory network inference.
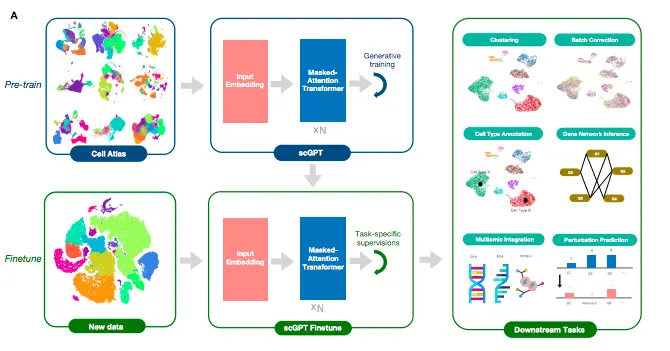
Image Source: https://doi.org/10.1101/2023.04.30.538439
State-of-the-art performance of scGPT
scGPT is capable of extracting features from previously unseen datasets. In the benchmarking experiments, scGPT outperformed recent methods and achieved state-of-the-art results across all downstream tasks.
- scGPT has demonstrated state-of-the-art performance in preserving the biological variance of the integrated datasets upon batch correction. scGPT successfully integrated all batches of CD4+ T-cells, CD8+ T-cells, and CD14+ T-cells from the Immune Human dataset.
- For the cell annotation task, the authors finetuned the scGPT model for the hPancreas dataset of human pancreas cells. The scGPT model generated predictions with 96.7% accuracy.
- For the perturbation prediction task, scGPT achieves the highest correlation for seven out of the eight metrics for perturbation prediction analysis.
- scGPT facilitates multi-omic integration as well as multi-modal representation learning.
- scGPT demonstrates its ability to group functionally related genes and differentiate functionally different genes from the embedding network.
Conclusion
The development of scGPT is a remarkable feat for the single-cell domain and the scientific community. The demonstrated efficacy and efficiency of the foundation model upon finetuning for several downstream tasks in the single-cell analysis pipeline clearly establishes it as the foundation model that was required. With scGPT, the requirement for developing task-specific methods trained on specific datasets will be reduced if not totally abandoned. The incorporation of special attention masking along with multiple layers of transformers at the core of the scGPT renders it the first-of-its-kind foundation model for single-cell biology. In future versions, the authors envision training the model on large-scale datasets with more diversity, including multi-omics, spatial-omics, and diseased conditions. By incorporating temporal data, the authors hope to devise causal discoveries. In-context instruction learning for single-cell data could also become a possibility. In other words, the authors aim to further develop scGPT as a model that understands different tasks in the zero-shot setting and does away with finetuning, a true game-changer for the single-cell community, which would be a gift from the NLG domain.
Article Source: Reference Paper | scGPT codebase: GitHub Link
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.