Google scientists have developed a novel aging clock based on retinal images using deep learning techniques called “eyeAge.” The biological clock is often said to vary from chronological age for an individual and has been previously studied extensively using predictive aging clocks based on biomarkers such as DNA methylation. These often require blood samples and are intrusive. However, eyeAge is a non-invasive aging clock that uses fundus retinal images. The authors trained deep-learning models on fundus images from the EyePACS dataset in order to predict an individual’s chronological age. The novel aging clock is more accurate than other aging clocks, and the GWAS reveals deeper insights into the prediction of age-related diseases.
It is All in the Eye: how retinal images can determine biological aging
Aging results in changes throughout the body of an individual, often causing diseases due to molecular and physiological changes. Determining specific markers of aging is crucial as each individual is subjected to specific genetic and environmental conditions. Scientists have been involved in identifying such markers and have developed several aging clocks based on biomarkers such as the DNA methylation-based epigenetic clock as well as the phenotypic aging clock (a combination of chronological age and nine biomarkers of mortality, phenoAge).
Retinal microvasculature is an indicator of the overall health of the circulatory system and the brain of an individual, as is evident from the ongoing research. Changes in the eye with aging are found to be related to several diseases, such as diabetic retinopathy and neurodegenerative diseases, such as Parkinson’s and Alzheimer’s disease. Early detection of systematic diseases such as chronic hypertension and tumors is possible by ophthalmologists examining an individual’s eyes. While quite fascinating, the fact that eyes are the windows to disease prediction is not surprising as subtle changes in the vascular system are first reflected in the smallest of blood vessels, and retinal capillaries are among the smallest.
Deep learning aided aging clock: eyeAge
Instruments often fail to detect subtle changes in the smallest of capillaries, such as in the retinal microvasculature. Detection of such changes requires algorithmic approaches based n deep learning techniques.
Fundus imaging has shown to be an effective non-invasive method of identifying eye disorders. Deep learning techniques have been employed earlier for the detection of diabetic retinopathy from retinal images. This resulted in highly accurate disease prediction and often delivered results surpassing that of human experts.
The authors thus used a deep learning approach in developing an aging clock from fundus retinal images. The authors use deep learning models for predicting the chronological age from longitudinal fundus images. This novel aging clock is termed eyeAge, and the deviation of this predicted age from the actual chronological age is termed “eyeAgeAccel,” which is used in the analysis for mortality and the GWAS.
The authors tested the model performance on datasets from the UKBiobank comprising almost 120,000 retinal images. The prediction of an individual’s chronological age and the actual chronological age are found to closely match with a Pearson correlation coefficient of 0.87.
The following figure illustrates the highly accurate prediction of eyeAge.
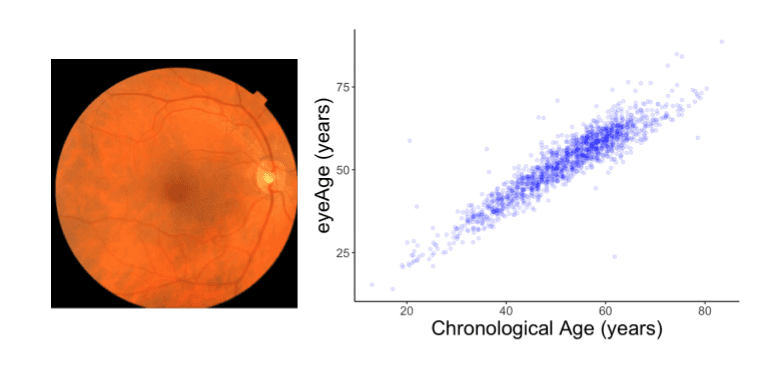
Image source: https://ai.googleblog.com/2023/04/developing-aging-clock-using-deep.html
eyeAge reveals deeper insights into biological aging
The authors found little correlation between the predicted chronological age from eyeAge and phenoAge, which is based on clinical blood test-based biomarkers. As was seen from the correlation figure for eyeAge and true chronological age, for certain individuals, the two are significantly different, thereby alluding to the fact that eyeAge is taking into account image features that are associated with diseases that are more prevalent in the aging population. They looked into it further and discovered that for conditions like COPD, myocardial infarction, and others, the predicted age is older than the real chronological age, indicating that the discrepancy is due to the existence of the disease.
The authors also performed a GWAS to identify genes associated with the difference eyeAgeAccel. Several of the hits were found to be associated with eye-related and age-related diseases. The knockdown experiments of the fly homolog Alk further validated the top hit in the GWAS as it was seen to slow the age-related vision decline in flies.
Conclusion
Researchers at Google have successfully developed a novel and highly accurate aging clock which predicts chronological age for an individual using longitudinal fundus retinal images based on a deep learning approach. The aging clock, eyeAge, is a never-before aging clock that has been based on retinal images and will be highly useful in predicting longevity. It will also have substantial applications in therapeutics as well as lifestyle assessment being non-invasive. The authors envision the use of eyeAge in future efforts toward developing anti-aging therapies. This is indeed a groundbreaking methodology for both the scientific as well as clinical communities.
Article Source: Reference Paper | Reference Article
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.










