Protein engineering is an emerging field in biological research – the improvement of existing proteins or the creation of new ones in order to improve efficiency or perform new functions has proved to be a field with much potential. Despite advancements in the field making protein engineering a sought-after field for industrial applications, it is still hampered by a lack of parameters by which different methods can be compared, a lack of large datasets that can be used as a basis for models, and a lack of accessibility for large numbers of researchers in the field. To rectify this, scientists from Align to Innovate, Cambridge, United States, and collaborators are introducing the Protein Engineering Tournament, a one-of-its-kind competition to characterize and design proteins in silico.
If cells can be considered living factories, proteins are the workers that allow life to persist by carrying out innumerable biochemical processes and functions. Composed of varying sequences of amino acids, these complex molecules do everything from ensuring structural stability to catalyzing chemical reactions. Their versatility and utility have made them the focus of much research in recent years. Protein engineering has emerged as a field with applications in nearly every area of biotechnology, from agricultural industries to therapeutics to environmental remediation. Engineered proteins can be used to recycle plastics, diagnose rare diseases, and reduce carbon emissions.
Advancements in technology have made the computational design of proteins an attractive prospect to many: theoretically, with the necessary inputs, a machine learning model should be able to train itself to recognize certain trends and mimic them when asked to make its own predictions. However, several things hamper its development: despite the creation of multiple machine learning models for this purpose, the scarcity of large, complex datasets, the lack of reproducibility, and the lack of available infrastructure to support the development and validation of such models make it difficult to take full advantage of the benefits that computational protein engineering has to offer.
The newly introduced Protein Engineering Tournament aims to tackle these drawbacks by serving as a platform where different methods can be benchmarked, suitable datasets can be generated, and open-source infrastructure can be developed. As it is conducted entirely online, the Tournament will eliminate geographic and financial barriers that many researchers may face. By providing a transparent platform through which protein engineering can be showcased and developed, the Tournament will encourage researchers to make their own forays into the field by providing the necessary infrastructure and support for research in the field to progress.
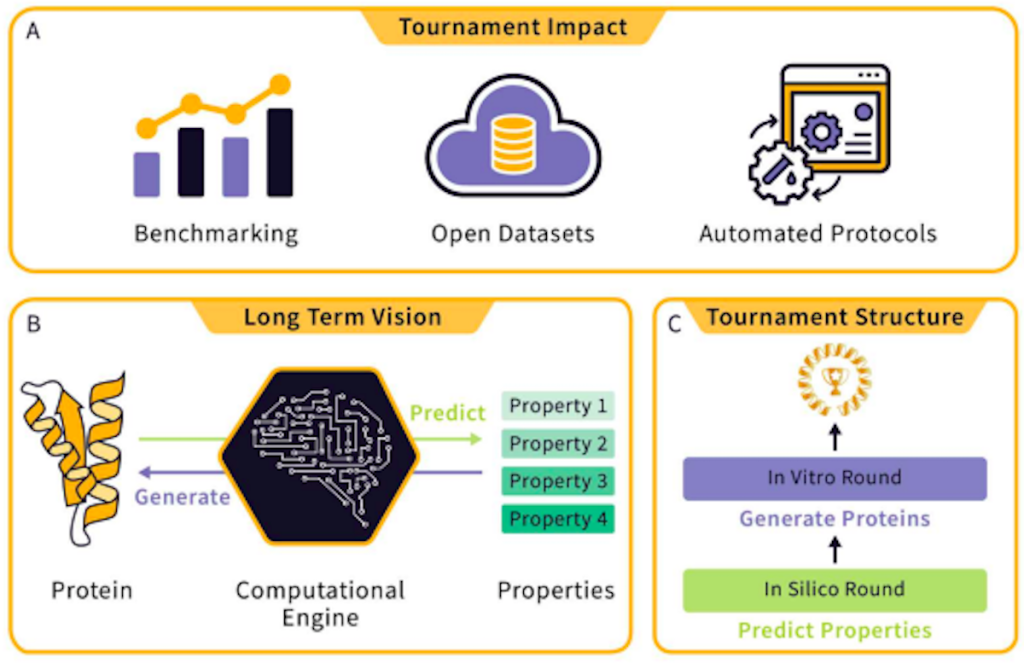
Image Description: Overview of the Tournament
Image Source: https://doi.org/10.48550/arXiv.2309.09955
The Tournament will also showcase the applicability of protein engineering to real-world problems by highlighting problems of significance in society that require solutions. This also includes problems that may, due to a lack of economic inducement, be ignored by industry and academia.
It has been demonstrated before that the availability of open datasets provides invaluable opportunities for researchers to develop and validate their methodologies. Previously, datasets such as ImageNet and MNIST provided labs with data to substantiate new methods while serving as a tool for measuring progress. Similar developments have also occurred in protein engineering, with the creation of tools like ProteinNet, TAPE, and FLIP. However, a lack of datasets of suitable size and complexity hinders collective progress in the area.
Competitions like the Protein Engineering Tournament will rectify this by providing researchers with novel datasets with which their methodologies can be tested and validated. For example, the Critical Assessment of Structure Prediction (CASP) has served as a mobilizing force in the field of protein structure prediction, allowing researchers to participate in a high-visibility event and motivating the development of new and improved methods. Such competitions can also serve as an entry point that is accessible to researchers outside academia, as demonstrated by DeepMind’s entry in the CASP competition in 2018. Similar competitions have spawned in CASP’s wake, dedicated to other fields, like CACHE, which focuses on computational chemistry.
The Protein Engineering Tournament aims to become a similar force for the community. It is comprised of two rounds: the first will be an in-silico round, and the second will be an in-vitro round.
The Protein Engineering Tournament – How It Will Be Conducted
The in silico round will consist of tasks where teams will be challenged with the development of models that can predict and infer various biophysical properties of proteins. Within this round, researchers can choose to directly predict such properties using protein sequence data (also known as zero-shot learning) or pre-training a model with an optional dataset (also known as supervised learning). Teams will be tasked with predicting the biophysical characteristics of various proteins based on their sequences. Such characteristics include stability, activity, and expressibility, among others. The submitted predictions will then be compared with experimental data to check for accuracy, using parameters like the Spearman correlation as a basis.
Different properties will be tested independently of each other, with entrants being allowed to submit their methods for whichever properties they choose. This allows labs that are building tools for the prediction of a specific property, like stability or enzymatic activity, to submit their tools for only those properties. After evaluation, the teams will be ranked, and a leaderboard will be made available, where the teams with the best performance will be allowed to progress to the second round. All datasets used in the competition will be published and will be available publicly for further research.
In the second round, teams will be tasked with designing protein sequences that satisfy or maximize given properties. The submitted sequences will be synthesized and characterized by the Tournament. The submissions will then be evaluated and ranked according to various parameters. After the conclusion of this round, all entrants will be ranked, with the highest-ranked team receiving the title of “Protein Engineering Champion.”
There are currently three avenues by which admission to the in vitro round will be considered: the first will be dependent on the teams’ performance in the in silico round. The second will allow researchers to apply directly to the in vitro round without needing to be in the preceding in silico round, allowing those with methodologies that aren’t suitable for the in silico round but which can be applicable in the in vitro round to participate. The third path to entry will require teams to submit a fee that will cover the experimentation expenses, eliminating the cost factor limiting the evaluation of all methods in the in vitro round.
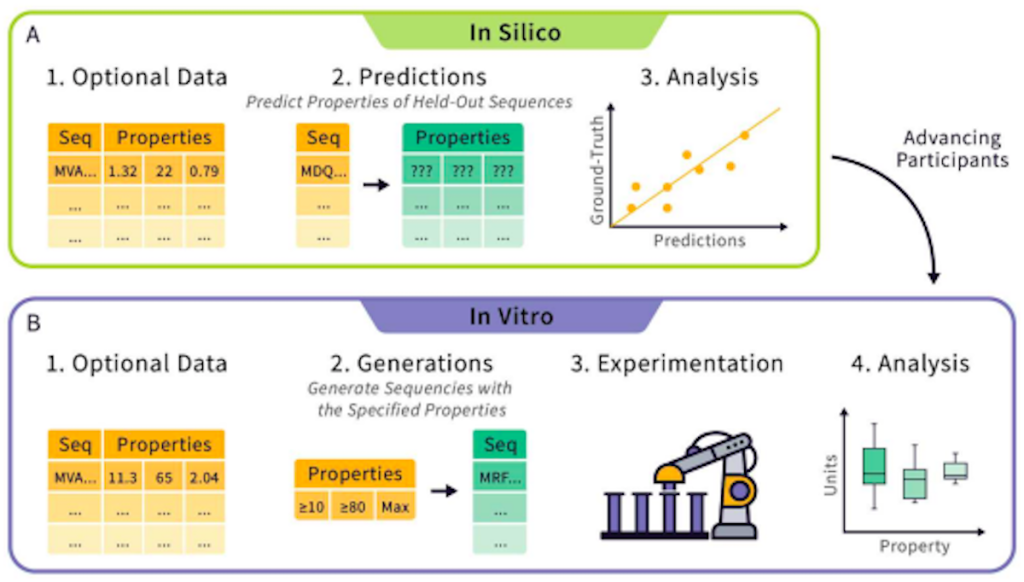
Image Source: https://doi.org/10.48550/arXiv.2309.09955
Conclusion
The tasks for the Tournament will be those that are considered societally important but may not be addressed by industry and academia due to a lack of necessary resources. The Tournament will also produce novel datasets for developing new tools, new assays for the characterization of proteins, and benchmarked results for the submitted tools. The Tournament will be designing and implementing its experimental workflows using cloud science labs that will allow the assays used to be accessible and reproducible to scientists. All data that is produced over the course of the Tournament will be made available publicly.
A pilot tournament has already been conducted, with Enzyme Design being the central theme. Six datasets were used, which were obtained from both academic and industry groups, with over 30 teams registered with varying levels of experience. It is hoped that the Tournament will act as an accelerating force in this regard by allowing researchers to benchmark models for the prediction of protein properties as well as the creation of new proteins that may have improved functioning in terms of efficacy and efficiency.
Article source: Reference Paper | Website
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Sonal Keni is a consulting scientific writing intern at CBIRT. She is pursuing a BTech in Biotechnology from the Manipal Institute of Technology. Her academic journey has been driven by a profound fascination for the intricate world of biology, and she is particularly drawn to computational biology and oncology. She also enjoys reading and painting in her free time.
















