The introduction of Full Laboratory Automations has served to revolutionize the workflows of labs worldwide, helping with the automation of routine tasks and the capacity for digital analysis of the results. However, a significant limitation is the inability to identify and interpret bacterial cultures, a task that is considered essential in the field of microbiology and diagnostics. A group of researchers from the University of Brescia seeks to remedy this with the introduction of a new deep-learning model, “DeepColony,” that can help with culture interpretation and quantification.
Despite the introduction of several new techniques in microbiology labs all over the world, the practice of bacterial culture and interpretation is still the primary method for diagnosing the presence of infectious pathogens. There are many reasons for its popularity, simplicity, and cost efficiency being prime among them. However, despite all of the advantages it offers, the interpretation of cultures accurately still remains a challenging task, prone to human error and constrained by the availability of qualified and experienced staff. Though other tasks requiring visual interpretation, such as cytology and hematology, have since been automated through the use of machine learning algorithms, bacterial culture remains unchanged for the most part.
Considering this lack of resources, the creation and development of Full Laboratory Automation systems have allowed for the automation and digitization of various steps involved in culturing, such as processing, streaking, and incubation, at the end of which images are uploaded for analysis. This has helped with the reduction of turn-around times, better efficiency and quality, as well as lower costs. However, the interpretation of these images still remains a significant challenge. While prior image analysis tools have been created for certain specific scenarios, these aren’t as helpful for other circumstances. The need for a generalized image analysis tool capable of consistent interpretations is great.
While similar solutions had been developed using conventional neural networks, the use of deep learning proved helpful in this case. The multi-network architecture was utilized due to its usefulness in complex decision-making algorithms. It was necessary that the information obtained from the algorithm be easy to procure and understand, necessitating the capacity to perform quantitation as well as identification of bacteria at a species level and be applicable for different kinds of culture analysis.
The Creation of DeepColony: A New Tool for Culture Interpretation
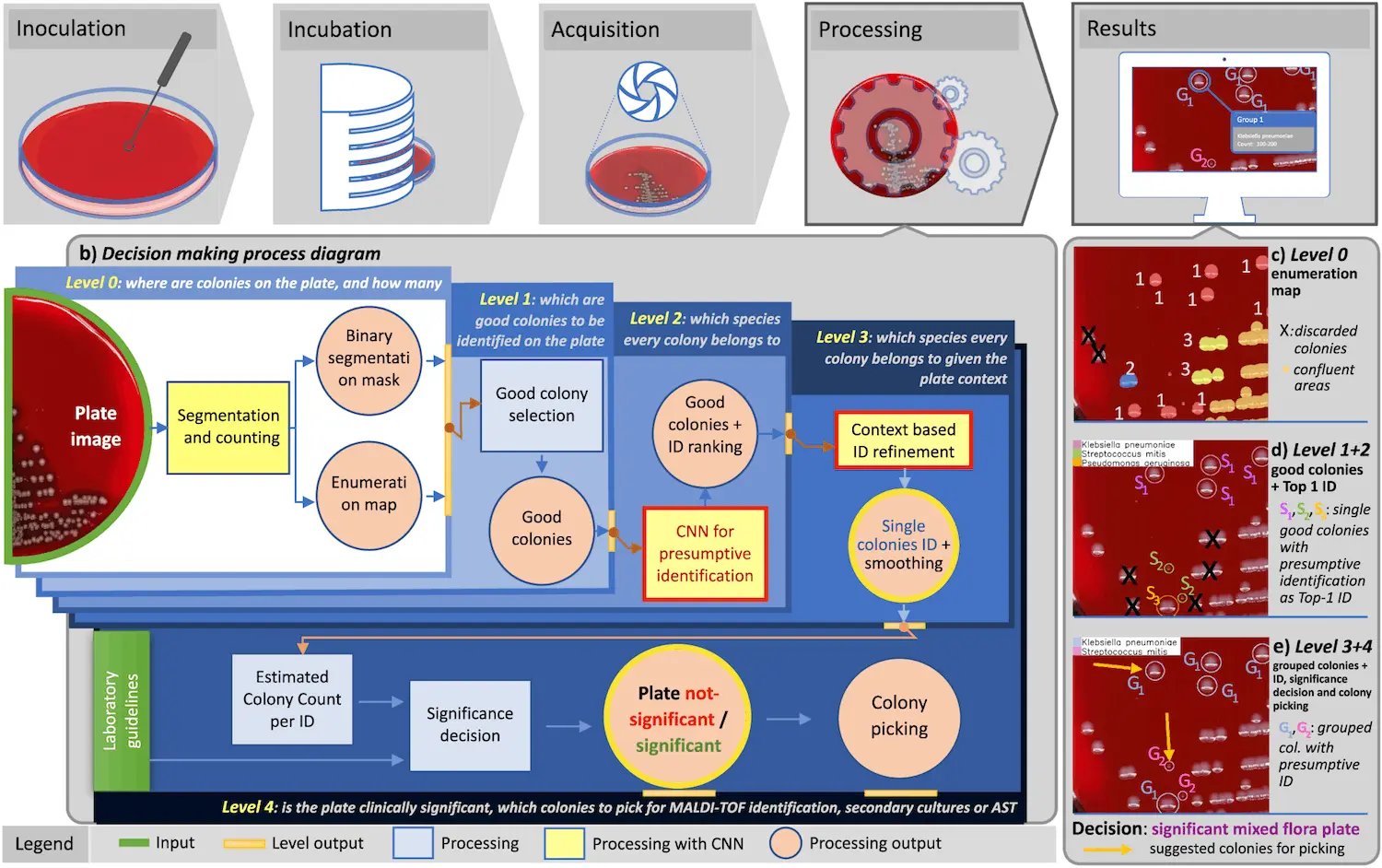
To address this need, DeepColony was created. DeepColony is a hierarchical multi-network that is able to perform various tasks for the identification, interpretation, and quantitation of cultures at different levels. Two methods are used in order to identify microbes at an accuracy level that is deemed suitable for clinical usefulness: the first being pathogen-aware – similarity agnostic and the second being similarity aware – pathogen agnostic. There are four different levels of analysis – individual colonies are assessed and identified at levels 0 – 2, and contextual information from surrounding colonies is taken into account at level 3. In order to classify colonies, human analysts take into account variations in the colony’s position, maturation, and dimensions. Using such data to train the machine learning model, it is possible to teach it to classify colonies using these same parameters. The information obtained is then used in the final level – level 4- in order to support decision-making.
Using more than 1300 unique images, a set of more than 26000 images was obtained of isolated colonies, which represented more than 32 species of bacteria and fungi that are commonly found in patients with urinary tract infections. Deep learning was used to create an algorithm that could accurately quantify bacterial growth using only images. Conventional neural network-based architecture was also utilized in order to obtain a list of possible identifications for a given colony, sorted by confidence in the prediction. The overall accuracy, when assessed after the bacteria were divided phylogenetically, was found to be 88.4%.
The information gained from the segmentation and enumeration of the colony is then used to create an interpretation of the culture. This information can be of great use to a laboratory. When applied to a dataset of more than 5000 urine cultures, DeepColony’s interpretations agreed with those of human analysts more than 95% of the time.
By providing methods by which the process of culture interpretation and diagnosis can be improved, namely in terms of scheduling, efficiency, and traceability, it is possible to lighten the workloads of employees and allow them to focus their time and skills on cases that require more effort or caution, resulting in greater productivity. This also reduces the need for manual interpretation, allowing for a reduction in costs that would otherwise be spent on labor. The capacity to provide decision support and quicken the process for diagnosis allows the algorithm to significantly improve precision and workflow as a whole, with the potential to make the process of diagnosing and responding to infection much faster, resulting in positive effects on patient care.
In addition, it has been noted that such systems that are based on artificial intelligence and machine learning must have an acceptable degree of transparency so as to ensure trustworthiness and comply with new healthcare regulations. This is in tandem with the responsibility to demonstrate the tangible and significant benefit of such mechanisms over conventional, human-driven methods. The transparency employed in the development of DeepColony, as well as the provisions for customizable configurations, help make the mechanism as optimal as possible for varying scenarios.
Conclusion
Currently, certain limitations exist – namely, DeepColony’s inability to identify species in a reliable manner when located inside confluent areas. While this doesn’t affect the algorithm’s consistency, it is similar to the limitations that exist even within human technologists’ attempts to interpret cultures that may be indistinguishable from each other or have a high degree of polymorphic variability.
However, the unique framework that DeepColony provides has the potential to revolutionize the workflows of microbiology laboratories by allowing for greater productivity, efficiency, and precision through its ability to accurately identify and interpret cultures and provide support for informed decision-making.
Article source: Reference Paper
Learn More:
Sonal Keni is a consulting scientific writing intern at CBIRT. She is pursuing a BTech in Biotechnology from the Manipal Institute of Technology. Her academic journey has been driven by a profound fascination for the intricate world of biology, and she is particularly drawn to computational biology and oncology. She also enjoys reading and painting in her free time.